
Rapid and Precise Gender Differentiation in Date Palms (Phoenix dactylifera L.) Using High-Resolution Melting Analysis
Nipitpong Sawongta and Maslin Osathanunkul*Published Date : May 29, 2025
DOI : https://doi.org/10.12982/NLSC.2025.046
Journal Issues : Number 3, July-September 2025
Abstract The date palm (Phoenix dactylifera L.) is a dioecious plant, meaning that individual trees are either male or female. Knowing the gender of date palms is essential for agriculturalists aiming to produce dates. High-Resolution Melting (HRM) analysis is a powerful molecular technique for identifying species, cultivars, and gender of living organisms. Therefore, this study aims to use the HRM analysis for sex discrimination of the date palm cultivar Deglet Nour. The research was conducted in two main phases, including (i) collecting nucleotide sequences from the 12th chromosome, specifically focusing on single nucleotide polymorphisms (SNPs) and gene regions that could serve as markers for gender identification; and (ii) performing both in silico and in vitro HRM analyses to accurately determine the gender of the date palms. Gender determination was based on differences in melting curve profiles and melting temperatures (Tm) between male and female samples. Among the primers tested, the TIF2 primer (targeting a gene region) emerged as the most effective for distinguishing gender. The melting curves generated by TIF2 primers were distinctly different for male and female trees, and their melting temperatures varied accordingly. These findings demonstrate that HRM analysis coupled with TIF2 primers is a powerful molecular technique for sex discrimination of the date palm cultivar Deglet Nour.
Keywords: Deglet Nour, Gene region, Melting curve, Melting temperature, SNPs
Funding: The authors are grateful for the research funding provided by the Chiang Mai University, Chiang Mai, Thailand.
Citation: Sawongta, N., and Osathanunkul, M. 2025. Rapid and precise gender differentiation in date palms (Phoenix dactylifera L.) using high-resolution melting analysis. Natural and Life Sciences Communications. 24(3): e2025046.
INTRODUCTION
Date palm (Phoenix dactylifera L.) is a long-lived monocot plant belonging to the Are-caceae (palm family), with origins in the Middle East and North Africa. The date palm is dioecious, with distinct male and female trees, and reaches reproductive maturity between four to seven years. The fruit, commonly referred to as the date, is rich in vitamins, fiber, and sugars, making it a valuable nutritional resource. Additionally, dates hold cultural and religious significance, particularly in Muslim communities (Al-Mahmoud et al., 2012). Moreover, date palms exhibit resilience to climate change, demonstrating heat tolerance and high yields. Their substantial market value has consequently increased interest in date palm cultivation in Thailand (Kanoeithip et al., 2015).
Like other dioecious plants, only female date palms produce the coveted edible fruit, making them significantly more sought after in cultivation. However, the male tree plays a critical role as well: it provides the pollen necessary for fertilizing female flowers. Remarkably, just one male tree can provide enough pollen for several female trees. The desirable ratio of male and female date palms is 1:10, respectively. This ratio boosts pollination efficiency, leading to higher yields and improved overall production (Intha and Chaiprasart, 2019). Therefore, the gender identification also prevents unnecessary planting of excess male trees, conserving valuable resources and maximizing orchard profitability.
Traditionally, the gender of date palms is identified by examining flower morphology. Nevertheless, when the date palm is still in the seeding stage, this approach cannot be applied. Accordingly, molecular techniques have emerged as a valuable solution, offering the ability to distinguish male and female plants at any developmental stage. In previous research, Restriction Fragment Length Polymorphism (RFLP), Random Amplification of Polymorphic DNA (RAPD), PCR base marker, Sex Determination regions of the Y chromosome (SRY) marker, and the combination of RAPD with Sequence Characterized Amplified Region (SCAR) markers were used for sex identification of date palm (El-Kharbotly et al., 1998; Fahad et al., 2018; Intha and Chaiprasart, 2019; Mohei et al., 2019; Ben-Abdallah et al., 2000). However, these methods can be time-consuming and complex, requiring multiple primers and gel electrophoresis, which is labor-intensive and offers limited sensitivity. To solve those limitations, this research proposes another molecular technique known as High-Resolution Melting (HRM) analysis that enhances sensitivity and simplifies the identification process.
HRM analysis is a powerful approach for quickly and accurately detecting differences in short DNA sequences. DNA fragments are first amplified using Real-time PCR. After that, the slightly increasing temperature unwinds double-strand DNA (dsDNA) to single-strand DNA (ssDNA). During this melting process, fluorescent dye detection slowly decreases, generating a melting curve of each amplicon fragment. The characteristics of each melting curve are used to identify species of living things, authenticate medical or food products, and gender discrimination in animals (Robertson et al., 2010; Madesis et al., 2012; Osathanunkul et al., 2015; Osathanunkul, 2018; Osathanunkul et al., 2018; Osathanunkul and Madesis, 2019; Gao et al., 2020; Osathanunkul et al., 2022). In the case of date palms, the point mutation of DNA sequences between both genders of the date palm is essential for gender identification through HRM analysis. For effective gender differentiation, understanding the date palm genome is essential. Previous studies have pinpointed linkage group 12 as a promising candidate for gender-specific markers due to its high density of single nucleotide polymorphisms (SNPs) relevant to gender (Mathew et al., 2015). Moreover, specific mutations associated with male sterility and female suppression on the 12th chromosome further support its role in gender determination (Torres et al., 2018; Torres et al., 2021). By focusing on this chromosome, HRM analysis can streamline the process of identifying male and female date palms, This facilitates the efficient selection and management of planting stock, ensuring the maintenance of the optimal male-to-female ratio (1:10), which is critical for maximizing pollination and crop yield.
Globally, there are numerous date palm cultivars, yet only a select few have gained popularity in Thailand include Barhee, Deglet Nour, KL1, and Medjool. Among these, Deglet Nour has become one of the most highly valued varieties across Europe and Asia, with impressive annual production exceeding 724,890 tons (Chouicha et al., 2014). Given its popularity and economic importance, this study focuses on using High-Resolution Melting (HRM) analysis to accurately identify the gender of Deglet Nour date palms.
MATERIAL AND METHODS
Plant materials and DNA extraction
The plant materials used in this study consist of sexually mature date palms of the Deglet Nour cultivar. The gender of these date palms was confirmed based on flower morphology, ensuring accurate classification prior to further analysis. Leaflet tips from date palms were collected, with younger provided superior DNA quality and yield. A total of 222 leaf samples (147 female and 75 male) were obtained from date palms across three provinces in Thailand: Chiang Mai, Phrae, and Phayao. Each sample was ground into a fine powder using liquid nitrogen, followed by DNA extraction using an adapted isolation method (Doyle and Doyle, 1990; Arif et al., 2010). Briefly, 100 mg of powdered sample was placed in a 1.5 mL microcentrifuge tube, where 1 mL of CTAB solution and 15 µL of proteinase K were added. The mixture was then incubated at 60°C for one hour. After incubation, the supernatant was carefully removed, mixed with chloroform, and centrifuged at 14,000 × g. The resulting supernatant was transferred to a fresh tube, precipitated with isopropanol, and dissolved in TE buffer. The DNA concentration and purity were assessed with a NanoDrop spec-trophotometer (Thermo Fisher Scientific, USA). Finally, all DNA samples were adjusted to a concentration of 120 ng/µL and stored at -20°C for future analyses.
Data mining for exploring a sex determinate region
To enhance the accuracy of gender identification in date palms using HRM analysis, two regions of the 12th chromosome, containing SNPs and a gene region in male and female date palms, were targeted to design primers for precise gender determination of the date palm.
SNPs region
All of the SNPs regions between male and female date palms in the 12th chromosome were reported (Mathew et al., 2014). In this research, the SNPs between two genders were only selected for analysis (Table S1). The nucleotide sequence with the highest number of SNPs was retrieved from the GenBank and DPMMD databases (Table 1) and analyzed using the ClustalW alignment method in MEGA11 to identify conserved and variable regions. Then, the sequence data were also used to investigate a suitable primer for sex segregation in date palms (Figure S1–S6).
Table 1. Nucleotide sequences of SNPs and gene regions in the 12th chromosome were retrieved from GenBank (NCBI) for date palm with an accession number.
|
Name |
Sequence type |
Accession number |
|
PDK_30s1202771 |
SNPs in 12th chromosome |
GL744456 |
|
dpS12X |
Chromosome X reference |
MH680977 |
|
dpS12Y |
Chromosome Y reference |
MH680966 |
|
dpS2X |
Chromosome X reference |
MH680998 |
|
dpB2Y |
Chromosome Y reference |
MH681002 |
|
MYB101 |
Gene |
XM_008777199 |
|
BAG (X1) |
Gene |
XM_008804193 |
|
BAG (X2) |
Gene |
XM_008804194 |
|
TIF2 (X1) |
Gene |
XM_008777479 |
|
TIF2 (X2) |
Gene |
XM_008777480 |
Gene region
Three genes of the 12th chromosome that are located in both male and female trees and have a high potential to distinguish the gender of date palm include MYB101, BAG, and TIF2 (Torres et al., 2018). The nucleotide sequence of those genes was extracted from GenBank (Table 1). Next, the ClustalW alignment was used to identify conserved and variable regions in nucleotide sequences for designing an appropriate gender identification primer (Figure S7–S9).
Primer design and optimization
In order to design primers that can be used to identify the gender of the date palm, the primer pairs need to follow three criteria: (i) forward and reverse primers must be able to bind with conserved sites of both genders of the date palm; (ii) amplified regions from different sexes of the date palm must have high variations; and (iii) amplicon size should be between 150 and 300 bp. Other properties of primer, such as melting temperature, secondary structure, and primer-dimer, were analyzed by the OligoAnalyzer™ Tool (https://www.idtdna.com/ pages/tools/oligoanalizer). The melting curve profile was predicted with the uMELT software (https://www.dna-utah.org/umelt/umelt.html). The primer pairs that can generate the most different melting curves between male and female date palms were selected for specific tests and annealing temperature optimization with PCR and gel electrophoresis.
High-Resolution Melting (HRM) analysis
The selected primers were used to amplify DNA fragments using Real-time PCR. Real-time PCR and HRM analysis were conducted in Rotor-Gene Q (Qiagen, Hiden, Germa-ny). The reaction mixture for the HRM analysis was executed in a total volume of 20 µL. This contains 4 µL of 5x HOT FIREPol® EvaGreen® HRM Mix, 1 µL of 120 ng/µl DNA, 0.2 µM of forward primer and 0.2 µM of reverse primer (Table 2). Each sample was prepared in triplicate, and negative controls were included in every set of PCR reactions to ensure the validity of the results. The reaction was activated with an initial denaturing step at 94°C for 15 min, followed by 35 cycles of 94°C for 30 sec, 54 to 60°C for 30 sec, and 72°C for 45 sec. Normalized melting curves were generated after the last extension step by increasing the temperature from 75 to 97°C at 0.1°C/s by the Rotor-Gene Q software (Qiagen). The characteristics of the melting curve and melting temperature were used to distinguish the gender of the date palm.
Table 2. Nine primer pairs were designed from SNPs and gene regions for gender identification of the date palms in this study.
|
Region |
Primer Name |
|
Primer (5’-3’) |
Tm |
Amplicon size (bp) |
Mismatch (bp) |
GAP (bp) |
|
SNPs |
SNP1 |
FW |
GAT GGT TAT CAG CTG GAT CAT CG |
55.3 |
323 |
7 |
9 |
|
RW |
ACA CAG AAG AAA CAT GTT CGC TC |
55.7 |
|||||
|
SNP2 |
FW |
GAA CAT GTT TCT TCT GTG TCC CAG |
56.0 |
322 |
17 |
5 |
|
|
RW |
CCG AGT CAA TTT GGG TAG CTG |
55.6 |
|||||
|
SNP3 |
FW |
GTA ATG CGA GGC TCT CTT GG |
55.6 |
198 |
17 |
0 |
|
|
RW |
CTC CCA ATC TCT TGC CAC AT |
54.9 |
|||||
|
SNP4 |
FW |
GAG ACT ATG GCC GAT GAT GC |
55.5 |
230 |
19 |
3 |
|
|
RW |
ACC CGA GAA ACC TCT CCA AT |
55.8 |
|||||
|
SNP5 |
FW |
CGA GAG GTG TTT GGA GTA TGA CTG |
57.0 |
234 |
21 |
0 |
|
|
RW |
GTG CTC AGG CTG CAG AGA TTA T |
57.1 |
|||||
|
SNP6 |
FW |
CCC ACA TTG CTT CGG CAG CTC C |
63.3 |
245 |
18 |
26 |
|
|
RW |
GCT CAC ACC GCA CGT AGG ACA GG |
63.9 |
|||||
|
Gene |
BAG |
FW |
GGA TCA TGA GGA GAA CGG GGT CGG |
62.9 |
207 |
0 |
12 |
|
RW |
GAG CTG ACG GAC ACC TCG TAC CT |
62.3 |
|||||
|
MYB |
FW |
CCC ACT TGG CAT CAG ACT CC |
58.0 |
237 |
6 |
0 |
|
|
RW |
CCT TCG ACC CAA ACG TTC AC |
58.4 |
|||||
|
TIF2 |
FW |
GCT CTA GAC CTA GTA GAC CAG TG |
54.8 |
261 |
6 |
3 |
|
|
RW |
GGC TTT GCT TGT AGC TTT GG |
54.8 |
RESULTS
Data mining and primer for gender discrimination
SNPs region
A total of 73 gender-specific SNPs between male and female date palms were identified on the 12th chromosome (Table S1) (Arif et al., 2010; Torres et al., 2018). Contig PDK_30s1202771 (GL744456), which contained the highest number of SNPs, was selected for alignment with dpS12X (MH680977) and dpS12Y (MH680966). Based on this alignment, six primer pairs targeting the SNP region were designed (Table 2). The variation in DNA sequences across the six amplicons was analyzed, and the results revealed that the SNP6 fragment exhibited more gaps and nucleotide mismatches compared to the other fragments. As a result, the SNP6 fragment showed great potential for accurately identifying the gender of date palms, making it a promising target for gender discrimination in this study.
Gene region
The variation of nucleotides sequence between male and female date palms resulted in distinct melting curve shapes and differences in melting temperatures. To explore these variations, three genes (MYB101, BAG, and TIF2) were aligned with dpS2X (MH680998) and dpB2Y (MH681002). From conserved regions within these sequences, three primers targeting gene regions on the 12th chromosome were designed (Table 2). The variation of each amplicon was analyzed and it was found that both MYB and TIF2 amplicons contain six mismatch nucleotides. while the BAG and TIF2 amplicons had gaps, with BAG showing 12 gaps and TIF2 showing three. These nucleotide mismatches and gaps were key factors contributing to the variation in melting curve profiles, supporting the effectiveness of HRM analysis for gender discrimination in date palms.
HRM analysis
HRM stimulation by uMELT software
All primer pairs were simulated with melting curves using the uMELT software. The results revealed that the amplicons generated from all SNP primers displayed distinct melting curve patterns between male and female trees (Figure 1a–1f). However, among them, the SNP6 primer stood out and was selected for further testing due to its high amplicon variation and the highest melting temperature observed (Table 2). Additionally, primers targeting specific gene regions showed promising results. For instance, primers designed for the BAG and TIF2 gene produced a clear and significant difference in melting curves between the genders (Figure 2a–2c).
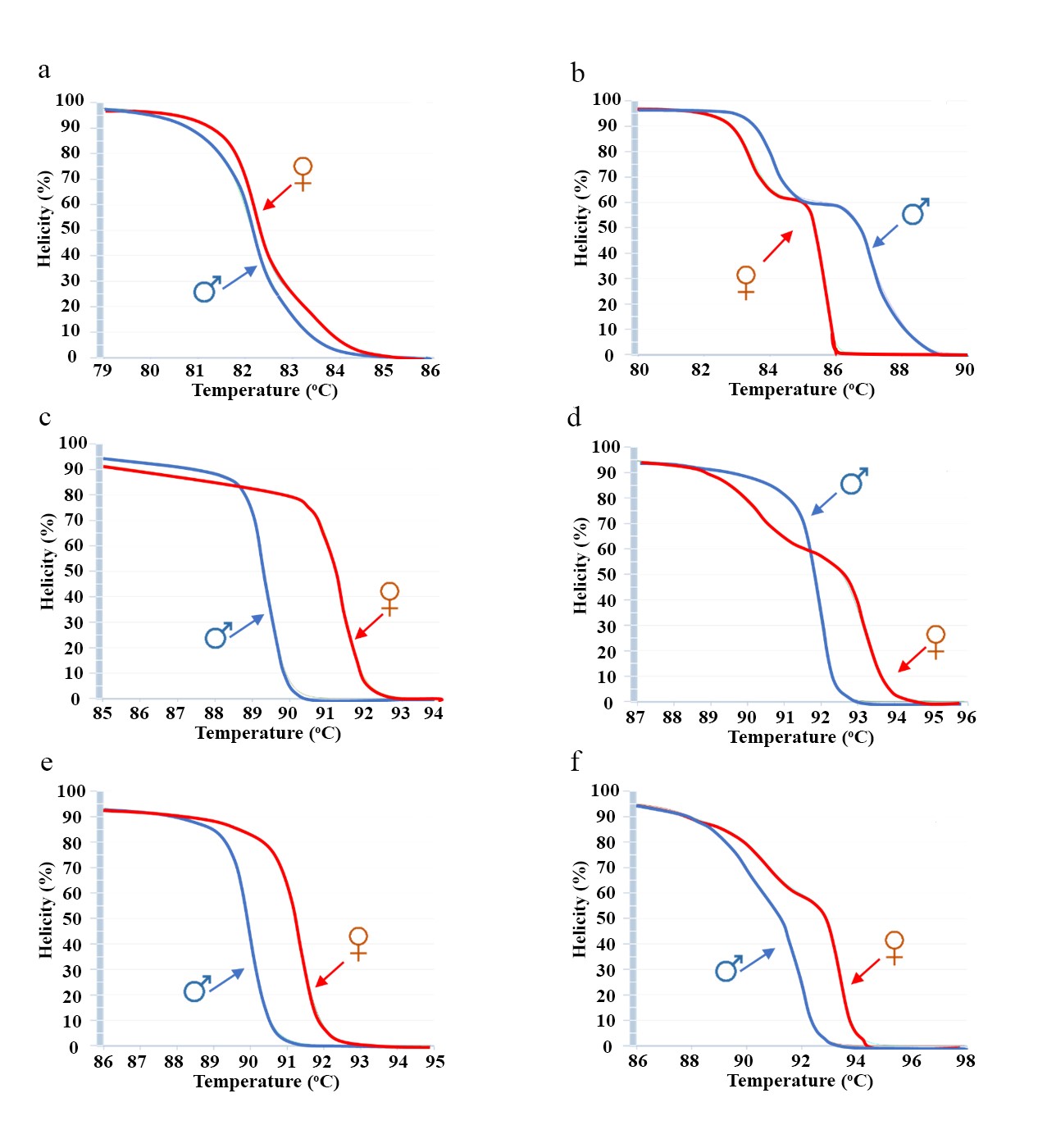
Figure 1. Melting curves analyzed by uMELT software in six SNPs primers include SNP1 primers (a), SNP2 primers (b), SNP3 primers (c), SNP4 primers (d), SNP5 primers(e), SNP6 primers (f).
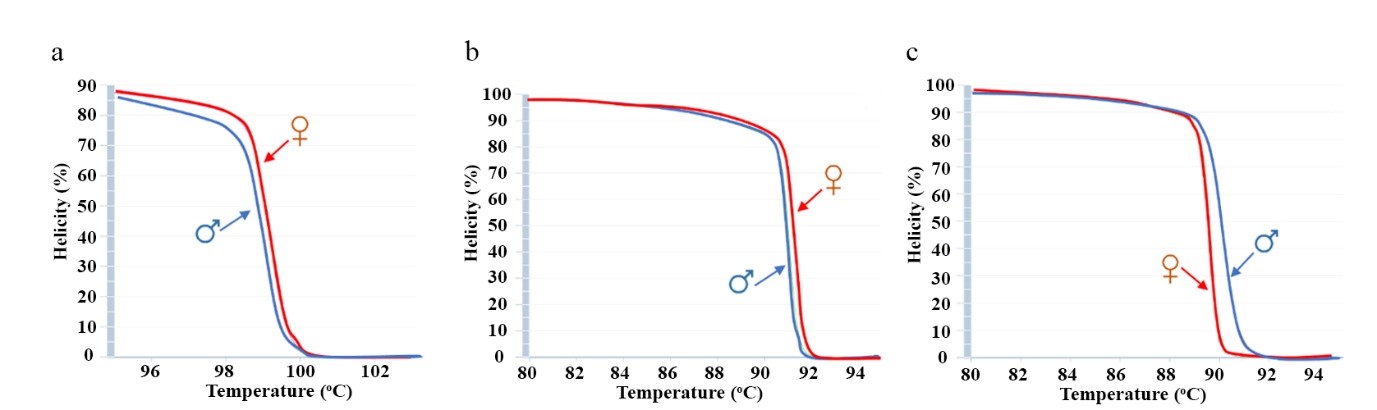
Figure 2. Melting curves analyzed by uMELT software in three gene primers include BAG primers (a), MYB primers (b), TIF2 primers (c).
in vitro HRM
The HRM analysis results for SNP6 (Figure 3a–3b), BAG (Figure 3c–3d), and TIF2 (Figure 3e–3f) primers with known sex samples revealed varying levels of effectiveness. While the SNP6 and BAG primers produced overlapping melting curves between male and female date palms, the TIF2 primers showed clear, distinguishable profiles. This unique ability to differentiate genders made the TIF2 primers ideal for further HRM analysis.
Using the TIF2 primers, the HRM analysis was conducted to determine the gender of date palm cultivar Deglet Nour across three Thai provinces (Chiang Mai, Phayao, and Phrae). The results indicated that male date palms exhibited a significant higher melting temperature of 86.87 ± 0.06 °C compared to female palms at 86.67 ± 0.06 °C (T-test, P < 0.05). Additionally, the melting curve profiles for male and female palms were notably distinct (Figure 4a–4f), underscoring the precision of TIF2 primers for gender differentiation in date palms.
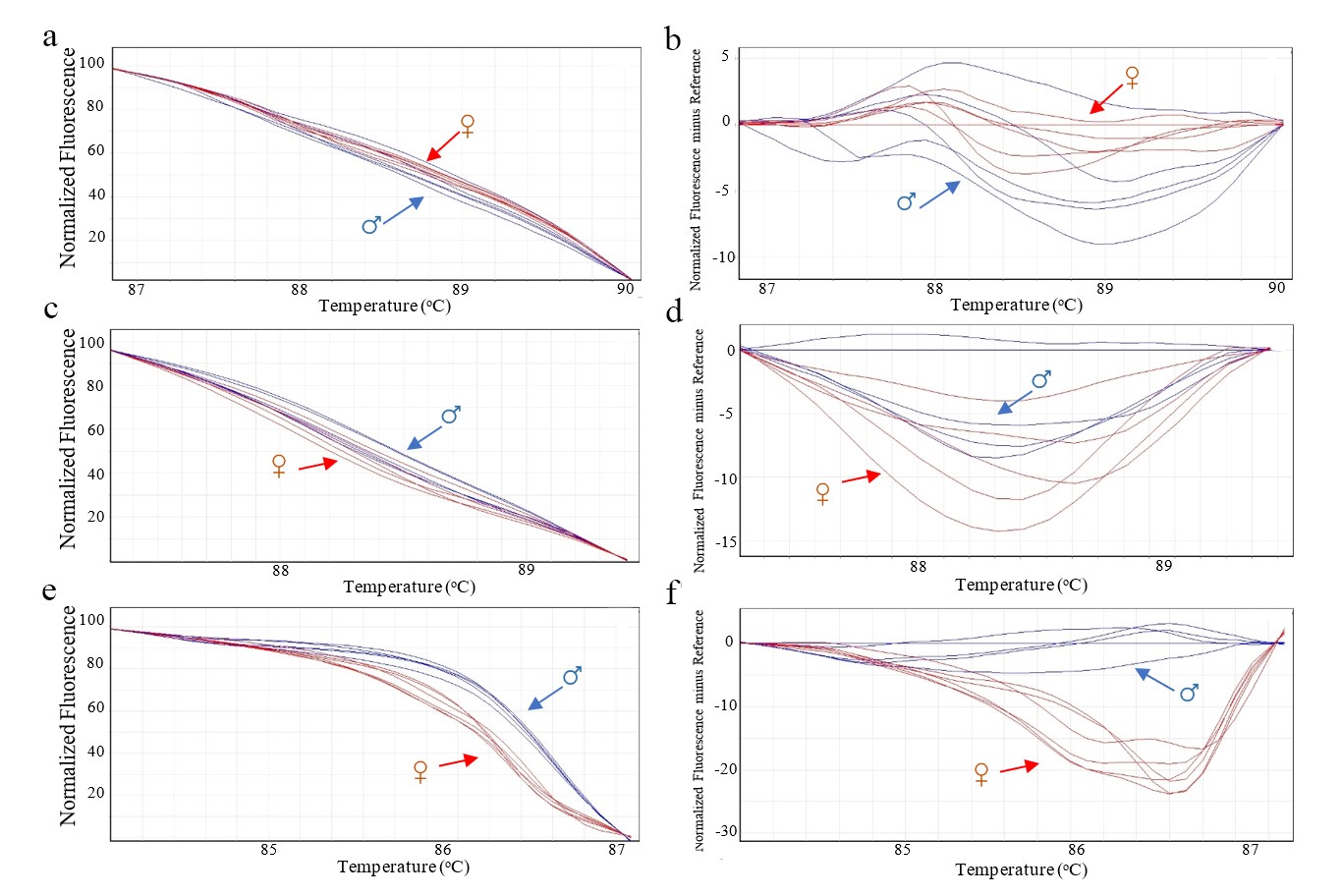
Figure 3. Melting curve profile of male and female date palm. Normalized fluorescence curve and male reference-corrected normalized fluorescence curve of SNP6 primers (a and b). Normalized fluorescence and male reference-corrected normalized fluorescence curve of BAG primers (c and d). Normalized fluorescence curve male reference-corrected normalized fluorescence curve of TIF2 primers (e and f). ♂: male date palm (Blue); ♀: female date palm (red).
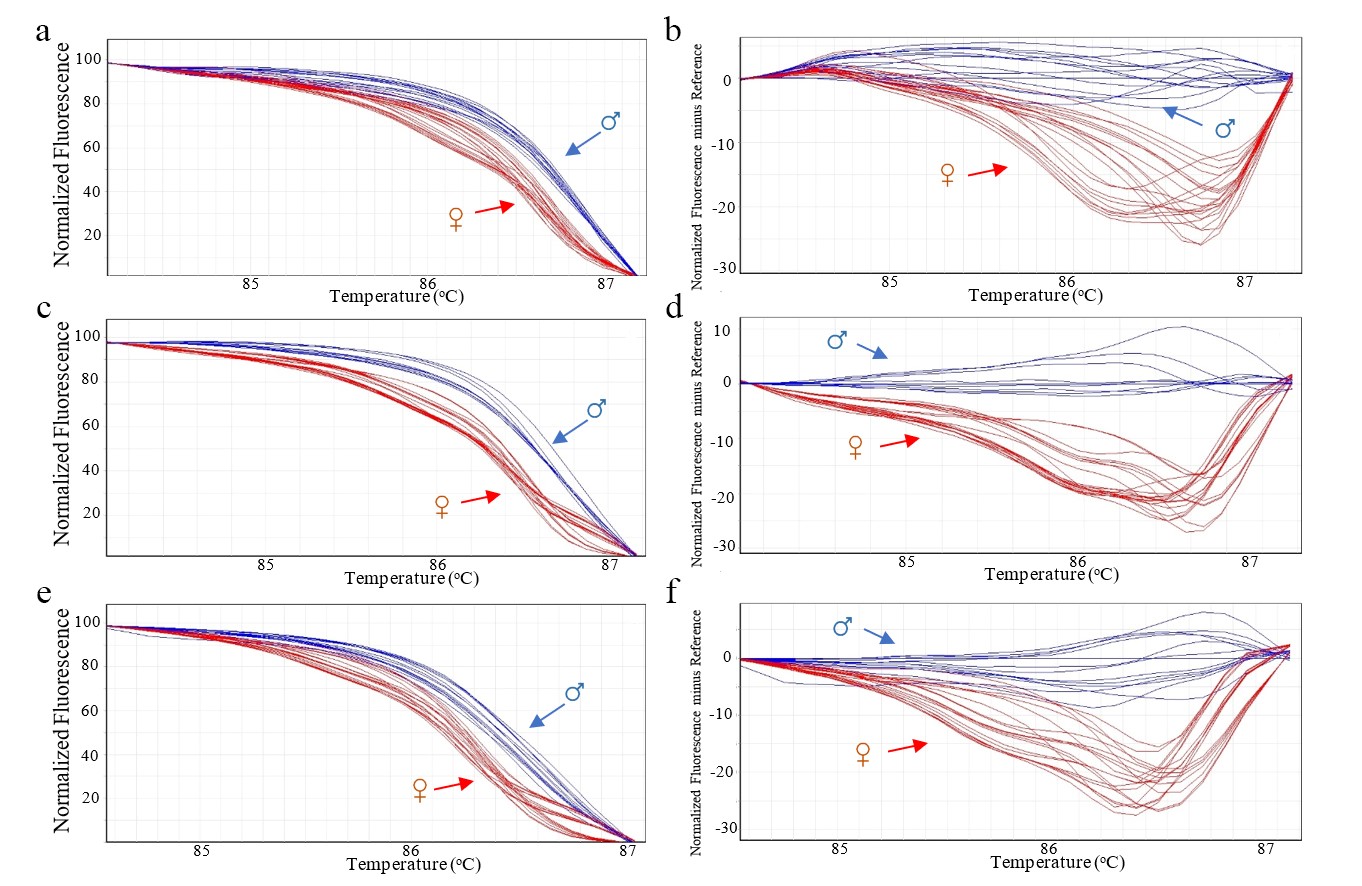
Figure 4. Melting curve profile of amplicons from TIF2 primers of male and female date palm in three provinces of Thailand. Normalized fluorescence curve and male reference-corrected normalized fluorescence curve of date palm in Chiang Mai province (a and b). Normalized fluorescence curve and male reference-corrected normalized fluorescence curve of date palm in Phayao province (c and d). Normalized fluorescence curve and male reference-corrected normalized fluorescence curve of date palm in Phare province (e and f). ♂: male date palm (Blue); ♀: female date palm (red).
DISCUSSION
HRM analysis is a fast and reliable technique widely used for gender identification across various animal species. This method has successfully been applied to discriminate gender in both terrestrial and aquatic animals, including Caribbean Flamingo (Chapman, 2011), chickens (England et al., 2021), Human fossil (Álvarez-Sandoval et al., 2014), Neotropical canids (include domestic dogs, wolves, and foxes) (González et al., 2015), pufferfish (Gao et al., 2020), and white leg shrimp (Perez-Enriqueza et al., 2019). In case of plant, HRM analysis has been effectively employed to determine the gender of cannabis (Gilchrist et al., 2021). Its efficiency in distinguishing gender makes HRM a valuable tool in diverse fields of biological research and agriculture.
The uMELT software has been widely utilized to assess the efficiency of primer pairs prior to performing HRM analysis (Xiu et al., 2020; Chen et al., 2021; Osathanunkul et al., 2022). However, in this study, uMELT's predicted melting curves did not match experimental HRM results. This discrepancy may result from GC content variability across sequences, leading to multiple melting domains (Dwight et al., 2011). Moreover, variables such as the thermodynamic parameter set applied during prediction and the type of fluorescent dye used in the experimental procedure have been shown to affect the shape and accuracy of melting curves (Dwight et al., 2012). Despite these limitations, uMelt remains a valuable and convenient tool for the initial design and optimization of HRM assays (Dwight et al., 2011).
Previous studies have shown that SNP loci combined with HRM analysis are powerful tools for species or cultivar identification in economic crops (Jeong et al., 2010; Ganopoulos et al., 2013). While HRM analysis with SNP regions has proven effective for gender identification in animals, this study demonstrates that SNP6 primers were unable to accurately identify the gender of the tested date palm samples. The presence of unpredicted mutations within the targeted fragments led to altered melting temperatures and melting curve characteristics, making the results unreliable (Wittwer, 2009). On the other hand, targeting gene regions, particularly conserved DNA regions, offers a more promising approach for gender identification in date palms. These gene regions are less likely to contain unpredictable mutations, making them more stable and reliable for gender classification. Furthermore, gene regions have been widely used for gender determination in animals, underscoring their potential for similar applications in plants (Morinha et al., 2011; Álvarez-Sandoval et al., 2014; Gao et al., 2020; England et al., 2021).
In terms of gene regions, BAG and TIF2 are commonly found in both dioecious plants (such as grape, melon, and papaya) and monoecious plants (such as citrus, peach, and pineapple). The BAG gene is involved in the regulation of protein folding, while TIF2 plays a role in the translation process. However, there is no conclusive evidence suggesting that these genes are directly related to gender determination. Despite their importance in protein function, neither BAG nor TIF2 has been widely used for gender identification in plants due to the lack of clear evidence regarding their specific role in sexual differentiation. Instead, male-specific regions such as CYP703, GPAT3, and APRT3 have been more commonly used for sex identification in plants (Torres et al., 2018; Massonnet et al., 2020; Leite Montalvão et al., 2021). However, these regions are not suitable for HRM analysis because they are exclusive to male plants. HRM analysis requires nucleotide sequence variation between male and female plants to generate distinct melting curves, and male-specific regions do not provide this variability in females.In this study, BAG and TIF2 were explored for the first time in relation to gender identification in date palms. The results showed that only the TIF2 primers were successful in distinguishing gender. The presence of nucleotide differences between male and female trees played a crucial role in this success, as mismatched nucleotides resulted in distinct melting curve profiles, enabling gender discrimination (Perez-Enriqueza et al., 2019; Keatley et al., 2020; England et al., 2021). In contrast, the BAG primers only detected a gap in the amplified fragment and did not exhibit any nucleotide mismatches between the male and female date palms (Table 2). This lack of nucleotide variation hindered the performance of the BAG primers in gender segregation through HRM analysis. Although the BAG gene is currently unsuitable for gender identification in date palms, it has shown potential in other areas, such as predicting breast cancer outcomes in humans (Papadakis et al., 2017), regulating heat stress and program of cell death in plants (Thanthrige et al., 2020; Arif et al., 2024), and its roles in various biological processes warrant further investigation in future studies.
HRM analysis is a powerful technique for detecting point mutations, and it has been widely used in various applications such as genetic testing (Andriantsoanirina et al., 2009) and identifying genetic variants linked to diseases (Rouleau et al., 2009). In this study, the fragment amplified by the TIF2 primer showed evidence of insertions/deletions (indels), missense mutations, and silent mutations due to nucleotide mismatches and gaps between the male and female date palms (Figure S10). In HRM analysis, point mutations are detected by the change in the melting behavior of the DNA fragment containing the mutation (Shih et al., 2009). These differences in melting behavior provide a clear and reliable method for detecting genetic variations, such as those associated with gender differentiation in date palms.
CONCLUSION
The date palm has emerged as a promising crop for Thai agriculturists. However, gender discrimination has traditionally taken four to seven years, making it crucial to develop a faster, more accurate method. This study used High-Resolution Melting (HRM) analysis to identify the gender of date palm (Deglet Nour cultivar). Primer pairs were designed from two regions of the 12th chromosome: six primers from the SNPs region (SNP1-SNP6) and three primers from the gene region (MYB, BAG, and TIF2). The results demonstrated that HRM analysis with the TIF2 primers successfully discriminated the gender of date palms cultivar Deglet Nour, offering a promising solution for quicker and more efficient gender identification.
ACKNOWLEDGEMENTS
This research was supported by Chiang Mai University. Moreover, we gratefully acknowledge Dailygreen farm, Kuntaphalum farm, and Wiangping farm for providing samples.
AUTHOR CONTRIBUTIONS
Nipitpong Sawongta contributed to the conceptualization, methodology, formal analysis, data visualization, and writing of the manuscript. Maslin Osathanunkul was responsible for the investigation, funding acquisition, project administration, resource provision, and contributed to the conceptualization, methodology, and writing of the manuscript. All authors have reviewed and approved the final version of the manuscript.
CONFLICT OF INTEREST
The authors declare that they hold no competing interests.
REFERENCES
Al-Mahmoud, M.E., Al-Dous, E.K., Al-Azwani, E.K., and Malek, J.A. 2012. DNA-based assays to distinguish date palm (Arecaceae) gender. American Journal of Botany. 99(1): e7-e10.
Álvarez-Sandoval, B.A., Manzanilla, L.R., and Montiel, R. 2014. Sex determination in highly fragmented human DNA by High-Resolution Melting (HRM) analysis. PLoS One. 9: e104629
Andriantsoanirina, V., Lascombes, V., Ratsimbasoa, A., Bouchier, C., Hoffman, J., Tichit, M., Rabarijaonab, L., Durande, R., and Ménard, D. 2009. Rapid detection of point mutations in Plasmodium falciparum genes associated with antimalarial drugs resistance by using high-resolution melting analysis. Journal of Microbiological Methods. 78(2): 165-170.
Arif, I.A., Mohammad, A.B., Haseeb, A.K., Anis, A., Ahmad, H.A., Farhan, A.H., Mohammad, A.S., Ali, H.B., and Mohammad, S. 2010. A simple method for DNA extraction from mature date palm leaves: Impact of sand grinding and composition of lysis buffer. International Journal of Molecular Sciences. 11: 3149-3157.
Arif, M., Men, S., Nawaz, A. F., Li, X., Xu, L., Yang, X., Fahad, S., Ahmad, P., Xu, R., and Li, L. 2024. Bcl-2-associated athanogene (BAG) Co-chaperones: Key players in multiple abiotic and biotic stress tolerance in plants. Journal of Plant Growth Regulation. 1-14.
Ben-Abdallah, A., Stiti, K., Lepoivre, P., and du Jardim, P. 2000. Date palm (Phoenix dactylifera L.) cultivar identification using random amplified polymorphic DNA (RAPD). Cahiers Agricultures. 9: 103-107.
Chapman, A. 2011. Development of novel high-resolution melting (HRM) assays for gender identification of Caribbean Flamingo (Phoenicopterus ruber ruber) and other birds. (Doctoral dissertation).
Chen, R., Chang, L., Cai, X., Wu, J., Liang, J., Lin, R., Song, Y., and Wang, X. 2021. Development of InDel markers for Brassica rapa based on a high-resolution melting curve. Horticultural Plant Journal. 7(1): 31-37.
Chouicha, S., Boubekri, A., Mennouche, D., Bouguetaia, H., Berrbeuh, M.H., Bouhafs, S., and Rezzoug, W. 2014. Valorization study of treated Deglet-nour dates by solar drying using three different solar driers. Energy Procedia. 50: 907-916.
Doyle, J.J., Doyle, J.L. 1990. Isolation of plant DNA from fresh tissue. Focus. 12: 13-15.
Dwight, Z., Palais, R., and Wittwer, C.T. 2011. uMELT: Prediction of high-resolution melting curves and dynamic melting profiles of PCR products in a rich web application. Bioinformatics. 27(7): 1019-1020.
Dwight, Z.L., Palais, R., and Wittwer, C.T. 2012. uAnalyze: Web-based high-resolution DNA melting analysis with comparison to thermodynamic predictions. IEEE/ACM Transactions on Computational Biology and Bioinformatics. 9(6): 1805-1811.
El-Kharbotly, A., El-Mardi, M., Al-Saadi, N., and Al-Mahruki, Y. 1998. Towards the construction of a genetic map of date palms using the amplified fragment length polymorphism technique (AFLP). p.194–207. In: Proceedings of the First International Conference on Date Palms, Al-Ain, United Arab Emirates, 8–10 Mar 1998. UAE University Press, Al-Ain
England, A.D., Kheravii, S.K., Musigwa, S., Kumar, A., Daneshmand, A., Sharma, N.K., Gharib-Naseri, K., and Wu, S.B. 2021. Sexing chickens (Gallus gallus domesticus) with high-resolution melting analysis using feather crude DNA. Poultry Science. 100: 100924.
Fahad, A.Q., Abdulhafed, A.A., Salim, K., Mohammad, N., Abdel-Rhman, Z.G., and Mohamed, T. 2018. SCAR marker for gender identification in date palm (Phoenix dactylifera L.) at the seedling stage. International Journal of Genomics. 3035406.
Ganopoulos, I., Tsaballa, A., Xanthopoulou, A., Madesis, P., Tsaftaris, A. 2013. Sweet cherry cultivar identification by High-Resolution Melting (HRM) analysis using gene-based SNP markers. Plant Molecular Biology Reporter. 31: 763-768.
Gao, F.X., Shi, Y., Duan, W., Lu, W.J., Huang, W., Zhang, X.J., Zhao, Z., Zhou, L., and Gui, J.F. 2020. A rapid and reliable method for identifying genetic sex in obscure pufferfish (Takifugu obscurus). Aquaculture. 519: 734749.
Gilchrist, E.J., Hegebarth, D., Wang, S., Quilichini, T.D., Sawler, J., Toh, S.Y., Foley, C., and Page, J.E. 2021. A rapid method for sex identification in Cannabis sativa using high resolution melt analysis. Botany. 101(7): 284-290.
González, S., Mannise, N., Repetto, L., and Maldonado, J.E. 2015. Sex determination of three Neotropical canids by High Resolution Melting analysis. Conservation. Genetics Resources. 7: 643-645.
Intha, N., and Chaiprasart, P. 2019. Sex determination in date palm (Phoenix dactylifera L.) by PCR based marker analysis. Scientia Horticulturae. 93: 251-255.
Jeong, H.J., Jo, Y.D., Park, S.W., and Kang, B.C. 2010. Identification of Capsicum species using SNP markers based on high resolution melting analysis. Genome. 53: 1029-1040.
Kanoeithip, C., Wilaiphon, S., Phomma, C., Ditkrachan, N., and Inthawong, S. 2015. Research and development of date palm. N.p.: Department of Agriculture, Bangkok (Thailand). Office of Agricultural Research and Development 1 (online access: https://agkb.lib.ku.ac.th/ doa/search_detail/result/385536)
Keatley, S., Botero, A., Fosu-Nyarko, J., Pallant, L., Northover, A., and Thompson, R.C.A. 2020. Species-level identification of trypanosomes infecting Australian wildlife by High-resolution melting - real time quantitative polymerase chain reaction (HRM-qPCR). The International Journal for Parasitology: Parasites and Wildlife. 19: 261-268.
Leite Montalvão, A.P., Kersten, B., Fladung, M., and Müller, N.A. 2021. The diversity and dynamics of sex determination in dioecious plants. Frontiers in Plant Science. 11: 580488.
Madesis, P., Ganopoulos, I., Anagnostis, A., and Tsaftaris, A. 2012. The application of Bar-HRM (Barcode DNA-High Resolution Melting) analysis for authenticity testing and quantitative detection of bean crops (Leguminosae) without prior DNA purification. Food Control. 25: 576-582.
Mathew, L.S., Spannagl, M., Al-Malki, A., George, B., Torres, M.F., Al-Dous, E.K., Al- Azwani, E.K., Hussein, E., Mathew, S., Mayer, K.F., Mohamoud, Y.A., Suhre, K., and Malek, J.A. 2014. A first genetic map of date palm (Phoenix dactylifera) reveals long-range genome structure conservation in the palms. BMC Genomics. 15: 285.
Massonnet, M., Cochetel, N., Minio, A., Vondras, A.M., Lin, J., Muyle, A., Garcia, J.F., Zhou, Y., Delledonne, M., Riaz, S., Figueroa-Balderas, R., Gaut, B.S., and Cantu, D. 2020. The genetic basis of sex determination in grapes. Nature Communications. 11: 2902.
Mohei, E.L., Mohasseb, H.A.A., Al-Khateeb, A.A., Al-Khateeb, S.A., Chowdhury, K., El-Shemy, H.A., and Aldaej, M.I. 2019. Identification and sequencing of Date-SRY Gene: A novel tool for sex determination of date palm (Phoenix dactylifera L.). Saudi Journal of Biological Sciences. 26(3): 514-523.
Morinha, F., Carvalho, M., Ferro, A., Guedes-Pinto, H., Rodrigues, R., and Bastos, E. 2011. Molecular sexing and analysis of CHD1-Z and CHD1-W sequence variations in wild common quail (Coturnix c. coturnix) and domesticated Japanese quail (Coturnix c. japonica). Journal of Genetics. 90: 39-43.
Osathanunkul, M, Madesis, P., and de Boer, H. 2015. Bar-HRM for authentication of plant-based medicines: Evaluation of three medicinal products derived from Acanthaceae species. PLoS One. 10: e0128476.
Osathanunkul, M. 2018 Bar-HRM for authenticating soursop (Annona muricata) tea. Scientific Reports. 8: 12666.
Osathanunkul, M., Osathanunkul, K., Wongwanakul, S., Osathanunkul, R., and Madesis, P. 2018. Multiuse of Bar-HRM for Ophiocordyceps sinensis identification and authentication. Scientific Reports. 8: 12770.
Osathanunkul, M., and Madesis, P. 2019. Bar-HRM: a reliable and fast method for species identification of ginseng (Panax ginseng, Panax notoginseng, Talinum paniculatum and Phytolacca Americana). PeerJ. 7: e7660.
Osathanunkul, M., Sawongta, N., Madesis, P., and Pheera, W. 2022. Bar-HRM for species confirmation of native plants used in forest restoration in Northern Thailand. Forests. 13: 997.
Papadakis, E.S., Reeves, T., Robson, N.H., Maishman, T., Packham, G., and Cutress, R.I. 2017. BAG-1 as a biomarker in early breast cancer prognosis: A systematic review with meta-analyses. British Journal of Cancer. 116(12): 1585-1594.
Perez-Enriqueza, R., Llera-Herrerab, R., Galindo, T.P., and Avila, S. 2019. A fast sex detection method for the white leg shrimp Litopenaeus vannamei by post-PCR high resolution melting (HRM). Aquaculture. 520: 734-784.
Robertson, T., Bibby, S., O’Rourke, D., Belfiore, T., Agnew-Crumpton, R., and Noormohammadi, A.H. 2010. Identification of chlamydial species in crocodiles and chickens by PCR-HRM curve analysis. Veterinary Microbiology. 145: 373-379.
Rouleau, E., Lefol, C., Bourdon, V., Coulet, F., Noguchi, T., Soubrier, F., Bièche, I., Olschwang, S., Sobol, H., and Lidereau, R. 2009. Quantitative PCR high‐resolution melting (qPCR‐HRM) curve analysis, a new approach to simultaneously screen point mutations and large rearrangements: Application to MLH1 germline mutations in Lynch syndrome. Human Mutation. 30(6): 867-875.
Shih, H.C., Er, T.K., Chang, T.J., Chang, Y.S., Liu, T.C., and Chang, J.G. 2009. Rapid identification of HBB gene mutations by high-resolution melting analysis. Clinical Biochemistry. 42(16-17): 1667-1676.
Thanthrige, N., Jain, S., Bhowmik, S. D., Ferguson, B. J., Kabbage, M., Mundree, S., and Williams, B. 2020. Centrality of BAGs in plant PCD, stress responses, and host defense. Trends in Plant Science. 25(11): 1131-1140.
Torres, M.F., Mathew, L.S., Ahmed, I., Al-Azwani, I.K., Krueger, R., Rivera-Nuñez, D., Mohamoud, Y.A., Clark, A.G., Suhre, K., and Malek, J.A. 2018. Genus-wide sequencing supports a two-locus model for sex- determination in Phoenix. Nature Communications. 9: 3969.
Torres, M.F., Mohamoud, Y.A., Younuskunju, S., Suhre, K., and Malek, J.A. 2021. Evidence of recombination suppression blocks on the Y chromosome of date palm (Phoenix dactylifera). Frontiers in Plant Science. 12: 634901.
Xiu, L., Li, Y., Wang, F., Zhang, C., Li, Y., Zeng, Y., Yin, Y., and Peng, J. 2020. Multiplex high-resolution melting assay for simultaneous identification of molecular markers associated with extended-spectrum cephalosporins and azithromycin resistance in Neisseria gonorrhoeae. The Journal of Molecular Diagnostics. 22(11): 1344-1355.
Wittwer, C.L. 2009. High-resolution DNA melting analysis: Advancements and limitations. Human Mutation. 30: 857-859.
OPEN access freely available online
Natural and Life Sciences Communications
Chiang Mai University, Thailand. https://cmuj.cmu.ac.th
Nipitpong Sawongta and Maslin Osathanunkul*
Department of Biology, Faculty of Science, Chiang Mai University, Chiang Mai 50200, Thailand.
Corresponding author: Maslin Osathanunkul, E-mail: maslin.cmu@gmail.com
Total Article Views
Editor: Tonapa Pusadee,
Chiang Mai University, Thailand
Article history:
Received: January 20, 2025;
Revised: April 24, 2025;
Accepted: April 29, 2025;
Online First: May 29, 2025

