
Evaluation of SNP-Based Markers Utilization for Resistance to Fall Armyworm Spodoptera frugiperda on Eight Corn Varieties
Paul Benyamin Timotiwu, Agustiansyah, Wawan Abdullah Setiawan, and Hamim Sudarsono*Published Date : July 10, 2023
DOI : https://doi.org/10.12982/NLSC.2023.039
Journal Issues : Number 3, July-September 2023
Abstract The aim of the study was to compare responses of different corn varieties to the fall armyworm Spodoptera frugiperda J.E. Smith (Lepidoptera: Noctuidae) attack and to map patterns of single nucleotide polymorphism of the RGA genes. The study consisted of two activities, i.e., the field experiment to observe corn damage intensity and the laboratory observation to study the SNP pattern. Field experiment was conducted to compare eight corn varieties set in randomized block design with 3 replications. Damage intensity of the corn due to S. frugiperda attack was observed on the field and the corn samples were analyzed in the laboratory to examine the RGA (resistance gen analogue) which contained single nucleotide polymorphism (SNP). The results from the field and the laboratory analysis showed that the response of Bisi 321 variety to S. frugiperda attack was better than the other tested varieties. Based on the electrophoresis of the PCR results with the SNP2_MNBS_Alt primer, samples of corn varieties coded as V1 (Pertiwi 5), V3 (P-36), V6 (NK Super), V7 (Exotic), and V8 (Local) produced amplicons with the size of ±150 bp. The samples that did not have amplification were V2 (NK7328), V4 (Bisi 321), and V5 (Bisi 18). There is a possibility that the amplification results may indicate some degrees of tolerance clusters of corn varieties to S. frugiperda attack. Further validation is required to apply the SNP-based markers of corn to develop tolerant or resistant varieties.
Keywords: Spodoptera frugiperda, RGA, SNP, Corn
Funding: This study was funded by the University of Lampung under the scheme of the Unila Professorship Research Grant 2021.
Citation: Timotiwu, P. B., Agustiansyah, Setiawan, W. A., and Sudarsono, H. 2023. Evaluation of SNP-based markers utilization for resistance to fall armyworm Spodoptera frugiperda on eight corn varieties. Natural and Life Sciences Communications. 22(3): e2023039.
INTRODUCTION
Since being reported in Lampung and other provinces in Indonesia, the fall armyworm Spodoptera frugiperda J. Smith (Lepidoptera: Noctuidae) has caused serious damage to corn crops. Morphological and molecular identification of collected larvae confirmed that the pest originating from the United States has entered the Lampung area (Sudarsono et al., 2019). The results of this identification further confirmed that populations of armyworm S. frugiperda found in Lampung were molecularly the same as those found in other countries that had experienced population explosions of this pest (Lestari et al., 2020). The fall armyworm S. frugiperda feeds on more than 80 plant species, including corn, rice, sorghum, sugarcane, vegetables and cotton. The larvae have voracious appetite, and they can multiply and spread quickly under the right environmental conditions (Abrahams et al., 2017).
Data from the Food Crops Protection Agency of Lampung Province showed that during the period of January to June of 2019, the damage caused by the fall armyworm S. frugiperda was recorded in 1,337 ha of corn field whereas the area of corn damaged by the local armyworm S. litura was only 93 ha. In other words, the damaged areas of corn caused by the invasive S. frugiperda was approximately 14 times higher than those caused by the local armyworms S. litura (Sudarsono, 2019)
Considering the serious threat posed by the fall armyworm, various measures must be prepared to anticipate the future outbreak of the pest. Utilization of resistant corn varieties is considered as one of the promising strategies to control the fall armyworm S. frugiperda. Therefore, it is necessary to select from various types of corn varieties grown in Indonesia to obtain resistant varieties or at least have good tolerance to the armyworm. Tolerant or resistant corn varieties are expected to produce profitable yield despite being attacked by the fall armyworm. The selected varieties can then be combined with other pest control strategies, including the chemical control strategy, to obtain a better control.
At present, there are at least six hybrid varieties of corns widely cultivated in Indonesia, especially in Lampung Province. Those corn varieties are: Pertiwi 5, NK7328, P36, Bisi 321, Bisi 18, and NK Super. In addition, there are also non-hybrid varieties of corn or local varieties cultivated by farmers in Lampung, i.e., the Exotic and the Local varieties. This field and laboratory experiments were conducted to evaluate and compare those eight varieties of corns for their resistance or tolerance to the fall armyworm attack. In general, plant resistance and tolerance to pests are influenced by the expression of resistance genes possessed by plants (Gao et al., 2006). One of the most important resistance genes is the resistance gene analogue (RGA) (Yaish et al., 2004). The relationship between the SNP pattern and its resistance to S. frugiperda attack could be evaluated by studying and comparing the pattern sequence of SNPs of the RGA gene of each corn variety. SNP patterns and performance of corn responses against S. frugiperda pests are important information in the development of resistant corn varieties.
Single nucleotide polymorphism (SNP) is a polymorphism caused by a single substitution process on nucleotides in the plant genome (Syvänen, 2001). Since the presence of SNPs is known to spread throughout the plant genome, SNPs have the potential to be used as molecular markers (Gupta et al., 2001). Considering that the utilization of SNP markers has been widely used in the study of plant diseases, there is also a possibility to use the markers for studying plant resistance or response to the fall armyworm attack or feeding. Accordingly, SNP diversity which is linkage with certain phenotypes has been reported in rice, soybean, and garlic (Gupta et al., 2001). Mutations due to one substitution can change the amino acid composition of the polypeptide encoded by the gene and can change the function of the polypeptide (Kowarsch et al., 2010). In order to use SNP markers for developing plant resistance or tolerance to pests, RGA-based SNP markers have to be evaluated for their polymorphisms and their relationship to the resistance properties of the pest. This study aimed to map different corn varieties response to S. frugiperda attack by comparing patterns of single nucleotide polymorphism (SNP) of the RGN genes.
MATERIALS AND METHODS
Planting of corn cultivars and field data collection
Field experiment was carried out in the corn producing region at Desa Margajaya, Metro Kibang District, East Lampung Regency, Lampung Province, Indonesia. The field experiment was run from May to October of 2021. This location was chosen because the population of the fall armyworm S. frugiperda was significantly high during the period of the study. Eight varieties of corn were planted on May 10, 2021 for this study: Pertiwi 5 (V1), NK7328 (V2), P36 (V3), Bisi 321 (V4), Bisi 18 (V5), NK Super (V6), Exotic (V7), and the Local variety (V8). These eight corn varieties were selected because they were most widely cultivated corn in the area. To provide equal opportunities for the feeding of the fall armyworm S. frugiperda, each tested variety was planted on an area of 2100 m2 (70 x 30 m).
Corn was planted with a spacing of 70 x 30 cm with a 70 cm of distance between rows. Each variety was planted in the same row so that the rows of plants in the experimental bed were alternately consisting of 8 tested varieties. Placement of rows of plants on the bed was done randomly. A sample of 5 plants located in each row of varieties was observed from each variety. Observation was conducted for each sample to measure the percentage of damage intensity of corn plants. Observation of the corn damage intensity due to attack by the fall armyworm S. frugiperda was carried out on leaves and cobs of corn. Leaf damage intensity was observed and rated using the criteria and scale developed by Davis et al. (1992) (Table 1). After each sample was observed and rated, damage percentage of the leaves was calculated with the following formula: Damage (%) for a corn plant leaf = [(total number of damaged leaves of single corn plant)/ (Total number of leaves assessed per single corn plant)] *100. Analysis of variance was performed on the data of corn leaf damage percentages, followed with Duncan’s test for the mean separation. R-Statistical (R Core Team, 2021) was used for the analysis of the data.
Table 1. Leaf damage criteria used to determine the damage intensity of corn varieties due to S. frugiperda attack (Davis et al., 1992).
|
Rating |
Explanation/definition of damage |
|
0 |
No visible leaf damage |
|
1 |
Only pin-hole damage |
|
2 |
Pin-hole and small circular hole damage to leaves |
|
3 |
Pinholes, small circular lesions and a few small elongated (rectangular shaped) lesions of up to 1.3 cm in length present on whorl and furl leaves. |
|
4 |
Several small to mid-sized 1.3 to 2.5 cm in length elongated lesions present on a few whorl and furl leaves |
|
5 |
Several large elongated lesions greater than 2.5 cm in length present on a few whorl and furl leaves and/or a few small- to mid-sized uniform to irregular shaped holes (basement membrane consumed) eaten from the whorl and/or furl leaves |
|
6 |
Several large elongated lesions present on several whorl and furl leaves and/or several large uniforms to irregular shaped holes eaten from furl and whorl leaves. |
|
7 |
Many elongated lesions of all sizes present on several whorl and furl leaves plus several large uniform to irregular shaped holes eaten from the whorl and furl leaves. |
|
8 |
Many elongated lesions of all sizes present on most whorl and furl leaves plus many mid- to large-sized uniform to irregular shaped holes eaten from the whorl and furl leaves. |
|
9 |
Whorl and furl leaves almost totally destroyed |
Genomic DNA extraction
The genomic DNA extraction and other laboratory analyses were conducted at the Integrated Laboratory and Technological Innovation Center, Lampung University. The following equipment were utilized for the genomic DNA extraction procedures: Dneasy Plan Mini Kit (50) (Qiagen, Cat#69104), Nanophotometer P 360 (Implen), QIAxcel Advanced, mortar, different size of tubes, vortex, heating block with 65°C of temperature, microcentrifuge, life touch 1.7 ml microcentrifuge tube, different sizes of micro pipettes, Bioclean Aerosol Resistant Barrier Tip, shaker, disposable free powder gloves 3.3.1, extract of genomic DNA samples taken from young leaf tissues of corn varieties using Dneasy Plan Mini Kit (50) (Qiagen, Cat#69104).
Fresh leaf samples of each corn variety weighing 100 mg were added with 400µL AP1, 4 µL Rnase A stock solution of 100 mg/mL. The mixture was then grounded with a sterile mortar before it was transferred to a 1.5 mL microtube for incubation in a 65°C heating block for 10 minutes. The tube was then shaken 2-3 times during incubation, added with 130 L Buffer P3, and incubated for 5 minutes. The lysate was centrifuged for 5 min at 20,000 x g before it was transferred to a QIA shredder column placed in a 2 ml collection tube for 2 minutes at 20,000 x g. The liquid was subsequently transferred into a new tube without disturbing the pellets, added with 1.5 volumes of AW1 buffer, and homogenized using a pippete. To remove the liquid, a total of 650 L of the mixture was transferred to a Dneasy Mini spin column placed in a 2 ml collection tube for 1 min at 6,000 x g. The spin column was placed into a new 2 ml collection tube, added with 500 L AW2 buffer, and centrifuged for 1 min at 6,000 x g. As much as 500 L AW2 Buffer was added and then centrifuged for 2 minutes at 20,000x g. The spin column was transferred to a new 1.5 ml or 2 ml tube and 100 L AE Buffer was added for elution. After incubated for 5 minutes at room temperature, the mixture again centrifuged for 1 min at 6,000 x g. The final DNA extract was stored in a freezer at -20°C.
DNA purity analysis and quantification
The analysis of DNA purity was carried out using a P360 Nanophotometer (Implen, Germany). After homogenized with a vortex, the sample purity was measured. The sample was pipetted and dropped in the middle of the measuring cell window and the sub-microliter cell containing the sample was tightly closed. The mixture was ready for DNA purity measurement. Following the completion of one sample reading, the lid and sub-microliter were cleaned using a moistened tissue before the measurement of the next sample.
After the completion of the DNA purity extraction, PCR (Polymerase Chain Reaction) test was conducted with the following primer pairs (Sutanto et al., 2016):
1) SNP1_MNBS_Ref,
2) SNP1_MNBS_Alt,
3) SNP2_MNBS_Ref,
4) SNP2_MNBS_Alt,
5) SNP4_MNBS_Ref,
6) SNP4_MNBS_Alt,
7) SNP5_MNBS_Ref,
8) SNP5_MNBS_Alt,
9) SNP6_MNBS_Ref,
10) SNP6_MNBS_Al0t
DNA amplification and electrophoresis
Thermocycler Sensoquest Sensodirect (SensoQuest, Gottingen, Germany) was utilized for the PCR DNA amplification. The first step of the procedure was to prepare the mixture of PCR reagents. PCR reaction was done at a total volume of 25 µL using KAPA2GTM PCR Kit. The composition of the PCR reagents was 5.0 µL 5 X PCR buffer; 0.5 µL 25 mM MgCl2; 0.5 µL 10 mM dNTPs; 1.0 µL 10µM for each primer (forward and reverse primers); 2.5 µL of genomic DNA (± 30 ng/µL); 0.1 µL Taq DNA polymerase (5 U/µL) and 15.4 µL ddH2O. The setting of the PCR equipment was done based on the method implemented by Sutanto et al. (2016). Initial denaturation was carried out at 95°C for 3 minutes, followed by 35 cycles of 95°C for 10 seconds, 60–62°C for 10 seconds, and 72°C for 3 seconds. After the cycle was completed, the procedure was ended with 1 cycle of 72°C for 10 minutes.
All PCR samples were electrophoresed using a QiaxCel an advanced digital electrophoresis apparatus from Qiagen, Germany using a DNA High Resolution Kit. The procedure was executed by following the QiaxCel Advanced manual.
RESULTS AND DISCUSSION
Percentage of corn damage intensity caused by S. frugiperda
Data of the fall armyworm attack on 5-week-old corn plants indicated that the plant damage intensity ranged from 49.8% to 61.3%. The lowest damage percentage was recorded on the Bisi 321 variety and the highest damage percentage was on the Local variety. Further analysis of variance of the data shows that there were significant differences of plant damage intensities among the tested varieties. At 5-week-old corn, damage percentage of the Local variety was significantly higher than those of Pertiwi 5 and Bisi 321. Damage level of the Local variety, however, was not significantly different from those of 5 other varieties (Exotic, NK7328, NK_Super, P36, Bisi_18) (Table 2). The boxplot of reordered data (Figure 1) confirmed the mean separation analysis in which the three lowest damage intensities were recorded on Bisi 321, Pertiwi 5, and Bisi 18, respectively.
Table 2. The average percentage of corn damage intensity at 5 weeks after planting due to Spodoptera frugiperda attack.
|
No. |
Code |
Variety |
Damage %* |
|
|
1 |
V8 |
Local |
61.33 ± 3.59 |
a |
|
2 |
V7 |
Exotic |
58.96 ± 3.54 |
ab |
|
3 |
V2 |
NK7328 |
55.11 ± 3.11 |
ab |
|
4 |
V6 |
NK_Super |
54.22 ± 3.56 |
ab |
|
5 |
V3 |
P36 |
52.30 ± 2.37 |
ab |
|
6 |
V5 |
Bisi_18 |
50.96 ± 2.15 |
ab |
|
7 |
V1 |
Pertiwi5 |
49.93 ± 4.77 |
b |
|
8 |
V4 |
Bisi_321 |
49.78 ± 1.33 |
b |
|
F-value = 5.11** |
||||
|
P-value = < 10-6 |
||||
Note: * Mean values followed by the same letter are not significantly different at 5% significance level with Duncan's test (DMRT).

Figure 1. Average of damage percentage of eight corn varieties due to S. frugiperda attack at 5 weeks after planting.
At seven weeks after planting, the corn growth was in the late vegetative phase before entering the flowering phase. Data of the fall armyworm attack on 7-week-old corn plants indicated that the plant damage intensities were only slightly higher than those on 5-week-old corn. The leaf damage intensities ranged from 50.8% to 62.7%. Consistent with the damage level on the 5-week-old corn, the lowest plant damage was recorded on Bisi 321variety and the highest plant damage level was recorded on the Local variety. Slightly different from the 5-week data, variety of Pertiwi 5 had lower damage intensity compared to Bisi 18 even though these were not statistically significant (Table 3, Figure 2). Overall, results of the analysis of variance of 7-week data also confirmed that there were significantly different responses of tested corn varieties toward S. frugiperda attack.
Table 3. The average percentage of corn damage intensity at 7 weeks after planting due to Spodoptera frugiperda attack.
|
No. |
Code |
Variety |
Damage %* |
|
|
1 |
V8 |
Local |
62.67 + 3.36 |
a |
|
2 |
V7 |
Exotic |
60.59 + 3.49 |
ab |
|
3 |
V3 |
P36 |
56.30 + 1.41 |
abc |
|
4 |
V2 |
NK7328 |
56.00 + 2.45 |
abc |
|
5 |
V6 |
NK_Super |
54.96 + 2.82 |
abc |
|
6 |
V5 |
Bisi_18 |
52.00 + 2.00 |
bc |
|
7 |
V1 |
Pertiwi5 |
51.85 + 1.93 |
bc |
|
8 |
V4 |
Bisi_321 |
50.82 + 0.97 |
c |
|
F-value = 15.93** |
||||
|
P-value = < 10-4 |
||||
Note: * Mean values followed by the same letter are not significantly different at 5% significance level with Duncan's test (DMRT).
Results of the Duncan’s test for 7-week after planting data indicates that variety of Bisi 321 had the lowest damage percentage due to the fall armyworm feeding even though it was not significantly different from those on Pertiwi5, Bisi_18, NK_Super, NK7328, and P36 varieties. Based on the results of the analysis, corn damage intensities due to the fall armyworm feeding in the study can be grouped into three categories, namely: 50-53% attacked (low), 54-56% attacked (medium), and > 57% (high) (Table 3). These categories may not directly indicate that variety Bisi 3221 was more tolerant or resistant than the other varieties since the study did not record the yield of the corn and the population of the fall armyworm was not monitored throughout the observation. The results, however, can be used as an early indicator of the relative response of the tested cultivars upon the fall armyworm attack. A more comprehensive study is required before the determination of the resistant or tolerant of corn variety.

Figure 2. Average of damage percentage of eight corn varieties due to S. frugiperda attack at 7 weeks after planting.
Analysis of SNP-RGA related to the resistance to S. frugiperda attack
Concentration and purity level of DNA extraction of the eight corn varieties are presented in Table 4. The electrophoresis image of the PCR results showed that only one primer pair produced PCR product, namely SNP2_MNBS_Alt. The other primer pairs of the PCR results did not produce PCR products (Figure 2). These results may imply that manipulation of the feature of single nucleotide polymorphism (SNP) on corn varieties could be used as the basis to develop varieties that have better response toward s. frugiperda attack. Single nucleotide polymorphism (SNP) is a polymorphism caused by a single substitution process on nucleotides in the plant genome (Syvänen, 2001). Since the presence of SNPs is known to spread throughout the plant genome, SNPs have the potential to be used as molecular markers (Gupta et al., 2001).
Table 4. Concentration and purity level of DNA extraction of the eight corn varieties.
|
No |
Varieties code |
Varieties name |
DNApurity (A260/A280) |
DNA concentration (ng/µL) |
|
1 |
V1 |
Pertiwi 5 |
1,978 |
3710 |
|
2 |
V2 |
NK7328 |
1,916 |
4540 |
|
3 |
V3 |
P36 |
1,953 |
3770 |
|
4 |
V4 |
Bisi 321 |
1,956 |
8800 |
|
5 |
V5 |
Bisi 18 |
1,938 |
2733 |
|
6 |
V6 |
NK Super |
1,934 |
3635 |
|
7 |
V7 |
Exotic |
1,897 |
3282 |
|
8 |
V8 |
Local |
1,954 |
2013 |
Data of the DNA purity and concentration indicated that all tested varieties in this study had high DNA purity. The highest DNA purity was recorded on the Pertiwi variety (1,978) and the lowest was observed on the Exotic variety (1,897). According to Sambrook and Russell (2001), DNA is determined as pure DNA if the purity value at the A260/A280 wavelength observation is between 1.8-2.0. In this study, the highest DNA concentration was obtained from the Bisi 321 variety (8,800 ng/µL) whereas the lowest concentration was recorded on the local variety (of 2013 ng/µL) (Table 4).
The electrophoresis results on Figure 3 indicated that V1, V3, V4, V5, V6, V7, and V8 samples had an amplicon band closest to the green band located at lower part of the chart (15bp size). The visible bands in these samples were identified as dimer, so they were not counted as amplicons (for the green band). Based on the electrophoresis of the PCR results with the SNP2_MNBS_Alt primer, the results were grouped into two clusters. The first cluster consisted of V1, V3, V6, V7, and V8; and the second cluster consisted of V2, V4, and V5.
Samples of corn varieties coded as V1 (Pertiwi 5), V3 (P-36), V6 (NK Super), V7 (Exotic), and V8 (Local) produced ±150 bp amplicons. The samples that did not have amplification were V2 (NK7328), V4 (Bisi 321), and V5 (Bisi 18). Based on these results, there is a possibility that the two clusters of amplification results may indicate some degree of more tolerant and less tolerant clusters of corn varieties to S. frugiperda attack. Further comprehensive validation is required to determine the possibility and the state of tolerance or resistance of corn varieties to S. frugiperda feeding.
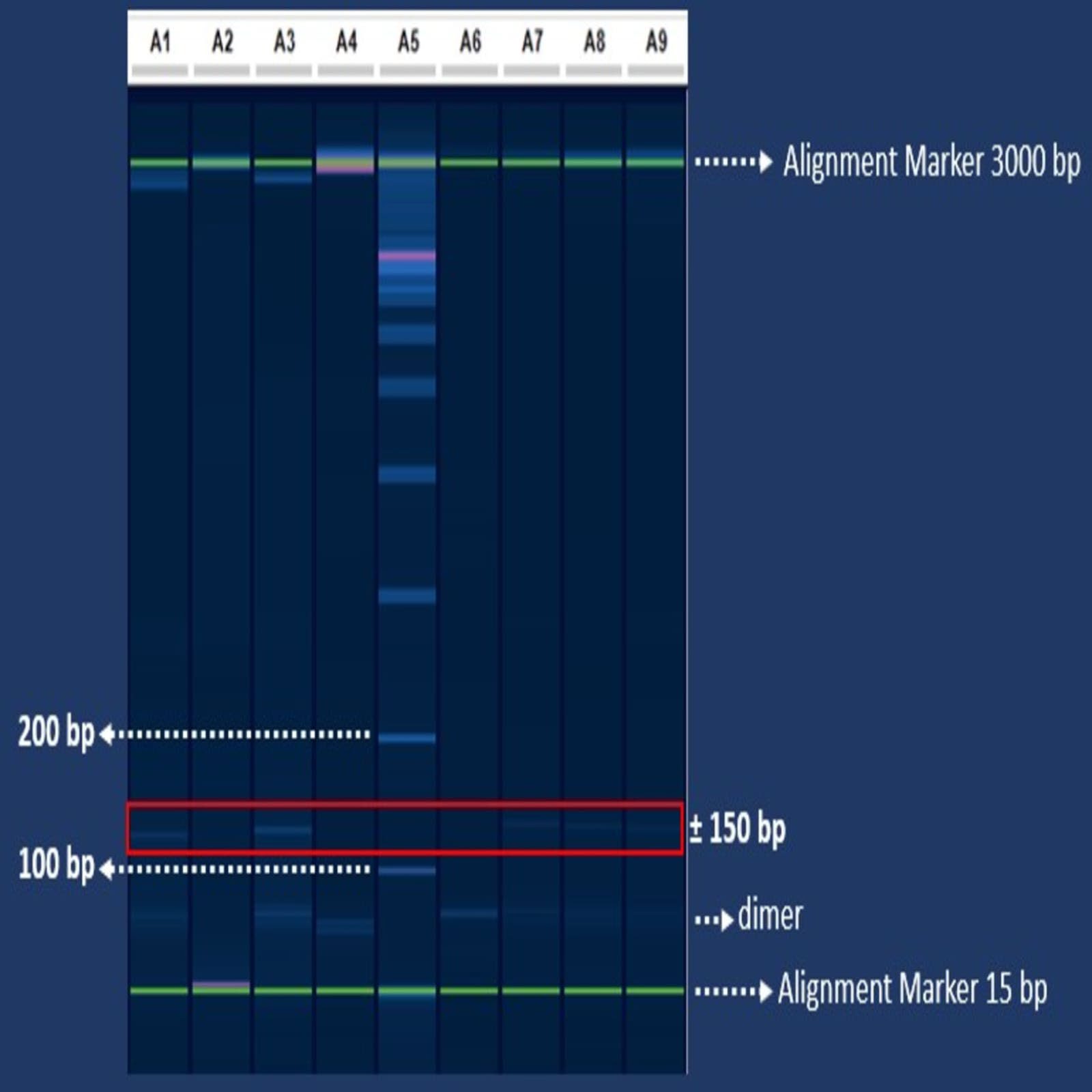
Figure 3. Electrophoresis results of RGA genes on 8 corn varieties. Notes: A1 = Var 1 (Pertiwi 50); A2 = Var 2 (NK7328); A3 = Var 3 (P36); A4 = Var 4 (Bisi 321); A5 = Size Marker; A6 = Var 5 (Bisi 18); A7 = Var 6 (NK Super); A8 = Var 7 (Exotic), A9 = Var 8 (Local). The two green bands are the QX Alignment Marker with 150 bp (below) and 3 kbp (above).
When compared to data from field observations, it is generally seen that varieties without amplicon (V2, V4, and V5) should have had lower damage rate (more tolerant) to the fall armyworm attack. However, field data showed that V4 (Bisi 321), V1 (Pertiwi 50), and V5 (Size Marker) were in the lower damaged group. Based on the electrophoresis data, V1 was in the cluster of medium or high damaged group whereas it ranks as the second low damaged group based on observation data in the field. This means that the data from electrophoresis were not able to distinguish between resistant and less resistant varieties. Therefore, a sequencing analysis is needed for the resulting amplicon. Through the results of the sequencing, the SNP of the corn genome RGA gene can be further determined to distinguish which varieties suffer higher damage due to fall armyworm feeding (less resistant to attack). In a more comprehensive study, therefore, SNPs and PCR setting can be potentially utilized to distinguish the status of tolerance or resistance of corn varieties toward the fall armyworm S. frugiperda attack.
CONCLUSION
The responses of eight tested corn varieties toward the fall armyworm S. frugiperda attack were grouped into three categories, i.e., 50-53%, (low), 54-56% (medium), and > 57% (high) plant damage intensity. Bisi 321variety was in the lowest damage group and it numerically has the lowest plant damage intensity at 5- and 7-weeks field observation. These results may be used as an early indicator of the relative response of the tested cultivars upon the fall armyworm attack.
Based on the electrophoresis of the PCR results with the SNP2_MNBS_Alt primer, samples of corn varieties coded as V1 (Pertiwi 5), V3 (P-36), V6 (NK Super), V7 (Exotic), and V8 (Local) produced amplicons with the size of ±150 bp. The samples that did not have amplification were V2 (NK7328), V4 (Bisi 321), and V5 (Bisi 18). There is a possibility that the amplification results may indicate some degree tolerant clusters of corn varieties to S. frugiperda attack. Further comprehensive validation is required to apply the SNP-based markers to determine the potential of tolerance or resistance of varieties toward S. frugiperda attack.
ACKNOWLEDGMENTS
This study was part of Professorship Research Grant 2021 funded by the University of Lampung. The authors thank the University of Lampung for providing the funding and facilities for this project.
AUTHOR CONTRIBUTIONS
Paul Timotiwu designed the overall study and performed the laboratory analysis at the Integrated Laboratory and Technological Innovation Center, Lampung University, and wrote the report. Hamim Sudarsono designed, conducted the field experiments, supervised the observation of corn damage intensity, performed the statistical analysis and data visualization, and revised the manuscript. Agustiansyah conducted the field experiment, data collection, and data analysis. Wawan A. Setiawan assisted in the laboratory work of genomic DNA extraction and SNP-RGA Analysis of SNP-RGA.
CONFLICT OF INTEREST
The authors declare that they hold no competing interests.
REFERENCES
Abrahams, P., Beale, T., Cock, M., Corniani, N., Day, R., Godwin, J., Murphy, S., Richards, G., and Vos, J. 2017. Fall armyworm status impacts and control options in Africa: Preliminary evidence note (April 2017).
Davis, F. M., Ng, S. S., and Williams, W. P. 1992. Visual rating scales for screening whorl-stage corn for resistance to fall armyworm. Technical bulletin - Mississippi Agricultural and Forestry Experiment Station (USA). 186: 9. http://europepmc.org/abstract/AGR/IND93032614
Gao, Y., Guo, W., Wang, L., and Zhang, T. 2006. Isolation and characterization of resistance and defense gene analogs in cotton (Gossypium barbadense L.). Science in China, Series C: Life Sciences. 49(6): 530-542.
Gupta, P. K., Roy, J. K., and Prasad, M. 2001. Single nucleotide polymorphisms: A new paradigm for molecular marker technology and DNA polymorphism detection with emphasis on their use in plants. Current Science. 80(4): 542-535.
Kowarsch, A., Fuchs, A., Frishman, D., and Pagel, P. 2010. Correlated mutations: A hallmark of phenotypic amino acid substitutions. PLoS Computational Biology, 6(9): e1000923.
Lestari, P., Budiarti, A., Fitriana, Y., Susilo, F., Swibawa, I. G., Sudarsono, H., Suharjo, R., Hariri, A. M., Purnomo, Nuryasin, Solikhin, Wibowo, L., Jumari, and Hartaman, M. 2020. Identification and genetic diversity of Spodoptera frugiperda in Lampung province, Indonesia. Biodiversitas. 21(4): 1670–1677.
R Core Team. 2021. R: language and environment for statistical computing. R Foundation for Statistical Computing.
Sambrook, J. and Russell, D. 2001. Molecular cloning: A laboratory manual, 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Sudarsono, H. 2019. Ulat Grayak Spodoptera frugiperda: Kondisi Terkini dan Analisis Dampaknya., Diskusi Roundtale CIPS Indonesia, 17 Juli 2019. Diskusi Roundtale CIPS Indonesia, 17 July 2019, Diskusi Roundtale CIPS Indonesia.
Sudarsono, H., Susilo F.X., Lestari P., Suharjo R., Swibawa I G., and Hariri A.M. 2019. Identification of Spodoptera Specimens Collected on Corn Field in Pringsewu District, Lampung Province. South East Asia Plant Protection Conference, 14 August 2019, Bogor. Indonesia.
Sutanto, A., Hermanto, C., Sukma, D., and Sudarsono, S. 2016. Pengembangan marka SNAP berbasis resistance gene analogue Pada Tanaman Pisang (Musa spp.). Journal Hortikultura. 23(4): 300.
Syvänen, A. C. 2001. Accessing genetic variation: genotyping single nucleotide polymorphisms. Nature Reviews. Genetics. 2(12): 930–942.
Yaish, M. W. F., Sáenz De Miera, L. E., and Pérez De La Vega, M. 2004. Isolation of a family of resistance gene analogue sequences of the nucleotide binding site (NBS) type from Lens species. Genome, 47(4): 650-659.
OPEN access freely available online
Natural and Life Sciences Communications
Chiang Mai University, Thailand. https://cmuj.cmu.ac.th
Paul Benyamin Timotiwu1, Agustiansyah1, Wawan Abdullah Setiawan2, and Hamim Sudarsono3, *
1 Department of Agronomy and Horticulture, Lampung University, Indonesia.
2 Department of Biology, Lampung University, Indonesia.
3 Department of Plant Protection, Lampung University, Indonesia.
Corresponding author: Hamim Sudarsono E-mail : hamim.sudarsono@fp.unila.ac.id
Total Article Views
Editor: Veerasak Punyapornwithaya,
Chiang Mai University, Thailand
Article history:
Received: February 24, 2023;
Revised: March 31, 2023;
Accepted: April 7, 2023;
Published online: April 21, 2023

