
Identification and Detection of a Virulence Gene of Streptomyces scabies Causing Potato Scab in Thailand
Angsana Akarapisan*, Athidtaya Kumvinit, Supaporn Falert and Wichai KositratanaPublished Date : April 3, 2023
DOI : https://doi.org/10.12982/NLSC.2023.027
Journal Issues : Number 2, April-June 2023
ABSTRACT
Potato scab is caused by several pathogenic Streptomyces species which diminish crop quality, quantity and marketability. In the present study, a 16S rRNA gene sequence was used to detect Streptomyces species in potato tuber lesions harvested in Chiang Mai, Thailand and to evaluate a virulence gene as a reliable marker for the detection of pathogenic Streptomyces species by PCR assays. Streptomyces isolates were isolated from potato scab lesions, of which one isolate was pathogenic on potato tubers. The pathogenic isolate MJ21 was identified as Streptomyces scabies based on 16S rRNA gene sequence and morphological characteristics. Subsequently, isolate MJ21 produced PCR products from the tomA and txtAB genes, which are related to the production of tomatinase enzyme and thaxtomin A, respectively. Moreover, when grown on nutrient agar (NA) with MJ21, eggplant seedlings showed severe stunting of the roots and shoots, and failed to germinate; by comparison, seedlings/seeds grown on NA plates without MJ21 exhibited no symptoms. This study reports that S. scabies MJ21 has a toxigenic region (TR) that is associated with the tomA and txtAB genes.
Keywords: Pathogenicity tests, Potato scab, Streptomyces scabies, Toxigenic region, Virulence gene
Citation: Akarapisan,A., Kumvinit,A., Falert,S., and Kositratana,W. 2023. Identification and detection of a virulence gene of Streptomyces scabies causing potato scab in Thailand. Nat. Life Sci. Commun. 22(2): e2023027.
INTRODUCTION
Domestic demand for potato has increased in recent decades in Thailand. (Kittipadakul et al., 2016). Potato scab disease of Solanum tuberosum is caused by several Streptomyces strains and is widely distributed over all the potato-growing areas in the world. The disease is characterized by superficial corky tissue or deep corky lesions or erumpent lesions on surface potato tubers, depending on the environment and the Streptomyces strain; slightly raised lesions may also occur. Although these symptoms have little or no effect on gross total yield, but they result in reduced starch content and lowered market value (Takeuchi et al., 1996). The main causal agent of common scab widely distributed is S. scabies, followed by S. turgidiscabies, S. luridiscabiei, S. acidiscabies and S. europaeiscabiei (Lerat et al., 2009). Several Streptomyces strains have been presented to infect potato tubers in North America and S. europaeiscabiei was more common in the west and S. scabies was the most common from the mid-plains eastward (Wanner, 2007). Moreover, the studies of Bouchek-Mechiche et al. (2000) and Park et al. (2003) suggest that Streptomyces species may have different population profiles in different parts of the world that cause common scabs. Streptomyces strains can cause potato scab disease worldwide such as S. scabiei, S. acidiscabies, S. europaeiscabiei, S. luridiscabiei, S. niveiscabiei, S. puniciscabiei, S. reticuliscabiei, S. stelliscabiei, S. turgidiscabies and S. ipomoeae (St-Onge et al., 2008; Atiq et al., 2013; Wanner and Kirk, 2015). Yang et al., (2018) reported that 10 different species of Streptomyces (S. scabies, S. turgidiscasies, S. acidiscabies, S. anulatus, S. europaeicabiei, S. enissocaesillies, S. luridiscaliei, S. caviscabies, S. aureofaciens, and S. griseus) found on potato common scab in Yunnan, China. Streptomyces isolates in China identified isolates primarily as S. scabies, S. acidiscabies, S. griseoflavus based on morphologic, biochemical and molecular analysis. Among the S. scabies showed the highest scab index by exhibiting the scab-like lesions on potato tubers (Ismail et al., 2020). The present study isolated Streptomyces strain from scabby potatoes in China shared sequence similarity with Streptomyces rhizophilus (Wei et al., 2022).
The pathogenicity island (PAI) is of importance for pathogenicity wherein the genes conferring pathogenicity are frequently clustered and appear to be responsible for the emergence of these more virulent strains or new pathogenic strains. These strains carry genes encoding virulence factors and four pathogenicities including a functional tomatinase (tomA) gene which virulence factor homologous to the gene encoding tomatinase, the biosynthetic pathway for thaxtomin (txt), a necrogenic protein (nec1) encodeing a protein that induces necrosis in plant tissue and a cytokinin biosynthetic pathway. These four virulence factors also exist in S. scabies, S. acidiscabies, S. turgidiscabies and some other Streptomyces strains, but are separated into two remote chromosomal regions, designated as the colonization region (CR) and the toxicogenic region (TR). Discovering the tomatinase homolog-encoding the tomA gene within the PAI of Streptomyces pathogens has led to speculation that this gene is also involved in virulence or pathogenicity in plant disease (Kers et al., 2005; Loria et al., 2006). In addition, the genes associated with thaxtomin production, such as the txtA and txtB genes called the ‘toxicogenic region’ are found in the first segment of the PAI. Whereas the virulence-related genes, nec1 and tomA, are located in the second segment, designated the ‘colonization region’ (Lerat et al., 2009).
The report in Thailand found that most Streptomycetes have characteristics as biocontrol and a few have characteristics as a plant pathogen. Shutsrirung et al. (2013) studied the diversity of endophytic actinomycetes in mandarin and their potential as plant growth promoters due to their ability to produce IAA. The isolates were classified into six genera including Streptomyces, Nocardia, Nocardiopsis, Spirillospora, Microbispora and Micromonospora based on spore chain morphology and 16S rRNA gene sequence. Whereas, Soe et al. (2010) reported that Streptomyces and Bradyrhizobia showed negative effect on nitrogen fixation in soybean. Similar results that Streptomyces strain RM365 was showed highest activity to inhibit bacterial pustule disease in the soybean strain but showed negative effect on the growth of Rhizobium, symbiotic bacteria of soybean plants. The strain shared 99.28% similarity to Streptomyces caeruleatus GIMN4T. These findings point out importance of checking the growth inhibitory effect of actinomycetes against rhizobia before being selected for potential biocontrol for field trial (Mingma et al., 2014.).
Careful and thorough characterization of Streptomyces pathogens and virulence related genes and their pathogenicity is a prerequisite for the development of better management procedures for controlling potato scab disease. The objectives of this research were to identify Streptomyces species and to evaluate the virulence genes for the detection of pathogenic Streptomyces species on potato tubers.
MATERIALS AND METHODS
Bacterial isolates
The bacterial isolates were obtained from potato tubers cvs. Atlantic and Spunta collected from Jom Thong District (18.2659, 98.6189), Muang District (18.7668, 98.9282) and Chai Prakan District (19.7534, 99.1515), Chiang Mai Province. Scab lesions were excised by scalpel (ca. 5 × 5 mm) and immersed in sodium hypochlorite solution (NaOCl) 1% (w/v) for 1 min and washed with sterile distilled water. After that, the potato pieces were placed on nutrient agar (NA), water agar (WA) and NA mixed with 150 μl of carbendazim at a concentration of 500 ppm (for fungal growth inhibition), and incubated at 25°C for 7 days (Lehtonen et al., 2004), after which colonies characteristic of Streptomyces were transferred onto fresh NA.
Pathogenicity assay
Pathogenicity tests were conducted on mini-tubers. Potato seedlings, cv. Atlantic, were firstly transplanted in the field and routine cultivation processes were performed thereafter. When the potato plants were close to flowering, stem cuttings were prepared and were transferred to pots containing sterilized sand, to produce mini-tubers. The isolates were grown on NA plates at 25°C for 7 days. Bacterial suspensions were prepared with sterile distilled water by serial dilution (1 x 108 CFU/ml). Then, 50 μl of the bacterial suspensions were inoculated to the mini-tubers and the tubers were recovered with sterilized sand. Inoculated plants were incubated at 25°C for 50 days (Miyajima et al., 1998).
Inoculation of Nicotiana tabacum was also performed to assess the pathogenicity of Streptomyces species using the protocol of Fyans et al. (2016). Leaves of 6-week-old N. tabacum plants were examined for necrosis in the area of infiltration.
Pathogenicity assay on plant seeds
Germination of eggplant seeds with Streptomyces sp. on NA plates and NA plates without bacteria used as controls was examined. The appearance of the seedlings was recorded after growth with the bacteria. When the seedlings showed hypertrophy and abnormal growth, or if the seeds did not germinate, the Streptomyces sp. isolates were considered pathogenic. The entire experiment was performed twice (Dees et al., 2013).
Identification of putative pathogenic Streptomyces by PCR
DNA was extracted using the protocol of Cheng and Jiang (2006). The virulence-related genes was determined by PCR using specific primers and PCR conditions for the tomatinase enzyme (tomA), thaxtomin A (txtAB) and the necrogenic protein (nec1) coding genes as previously described by Alejo et al. (2019). Primers and the expected sizes of the PCR products are indicated in Table 1. The PCR products were purified and directly sequenced. PCR products were sequenced and analyzed using a BLASTx algorithm-based program of MEGA10 and deposited in the GenBank database (Atiq et al., 2013).
Table 1. Primers used for PCR detection of virulence-related genes.
|
Gene |
Primer pair |
Product size (pb) |
|
nec1 |
Nf: 5’-ATGAGCGCGAACGGAAGCCCCGGA-3’ Nr: 5’-GCAGGTCGTCACGAAGGATCG-3’ |
700 |
|
txtAB |
TxtAB1: 5’-CCACCAGGACCTGCTCTTC-3’ TxtAB2: 5’-TCGAGTGGACCTCACAGATG-3’ |
385 |
|
tomA |
Tom3: 5’-GAGGCGTTGGTGGAGTTCTA-3’ Tom4: 5’TTGGGGTTGTACTCCTCGTC-3’ |
392 |
16S rRNA gene amplification
Amplification of the 16S rRNA gene sequence was performed using universal primers 16SF and 16SR for amplification of a 1,500 bp region of the 16S rRNA gene (Song et al., 2011). The PCR mixture was prepared in a total volume of 25 μl: containing 50-100 ng per μl of DNA, using Quick Taq HS DyeMix (TOYOBO CO., LTD.) and 20 μM of each primer. PCR conditions were as follows: 5 min at 94°C, followed by 30 cycles consisting of 30 sec at 94°C, 64°C for 30 sec, 72°C for 1.5 min and followed by a final extension at 72°C for 10 min (Lehtonen et al., 2004). The PCR products were purified and directly sequenced. The sequences of 16S rRNA genes determined in this study were aligned with the Clustal W program. An evolutionary tree was inferred using the neighbour-joining method of MEGA10 and assessed by performing bootstrap analysis on 1,000 resamplings (Song et al., 2004).
RESULTS
Bacterial isolates
In this study, four isolates of Streptomyces species were isolated from necrotic lesions of diseased potato tubers collected in Chiang Mai Province including Jom Thong District (MJ20 and MJ21), Muang District (MM03) and Chai Prakan District (PE04). On NA, Streptomyces spp. isolates MJ20, MJ21, and MM03 mycelia were sympodially branched and formed flexuous spore chains containing 20 or more spores which rarely curled at the tip as similarly report by Lambert and Loria (1989). The spores were smooth and cylindrical. The spores of all isolates were white (Figure 1a-c). In addition, the aerial mycelium of Streptomyces isolate PE04 was sympodially branched and formed spiral spore chains containing 20 or more spores which rarely curled at the tip. Spores were smooth and cylindrical. The spores of all isolates were white but turned to grey after 5 days (Figure 1d).
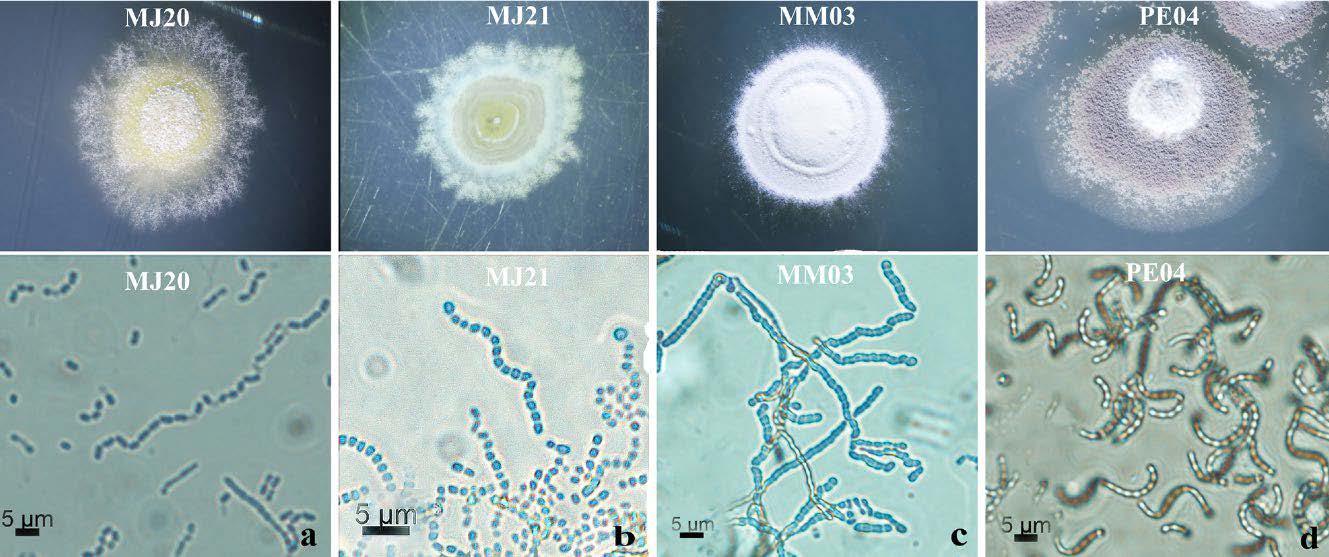
Figure 1. Cultural and micro-morphological characteristics of Streptomyces isolates: Colonies in nutrient agar at 28°C (top). Micromorphology of aerial growth appears as white hyphae under light microscope 100x (below) (Bar = 5 μm), (a) Streptomyces isolate MJ20, (b) Streptomyces isolate MJ21, (c) Streptomyces isolate MM03, (d) Streptomyces isolate PE04.
Pathogenicity assay
In pathogenicity tests, Streptomyces sp. isolate MJ21 induced raised or deeply-pitted corky lesions when compared with control under incubator conditions (Figure 2). Similar to scab lesions on field-grown tubers; but isolates MJ20, MM30 and PE04 did not produce lesions during the mini-tuber assay. In addition, a N. tabacum leaf infiltration assay showed that only isolate MJ21 could induce rapid necrosis in the area of infiltration. This later observation is of particular interest as it suggests that another virulence factor besides a phytotoxin. The isolate MJ21 is responsible for the observed tissue necrosis.

Figure 2. Pathogenicity of Streptomyces isolate MJ21on potato, (a) control, (b) symptoms on infected potato mini-tubers.
Pathogenicity assay on plant seeds
In pathogenicity tests, one of the putative pathogenic Streptomyces isolate MJ21 was tested on eggplant seeds. The abilities of isolate MJ21 to inhibit eggplant seeds germination were consistent with the presence of the phytotoxin, the pathogenicity determinant. The eggplant seeds grown with MJ21 failed to germinate. Although, the seeds grown on NA plates without bacteria germinated normally (Figure 3). Therefore, MJ21 can produce a phytotoxic secondary metabolite that inhibited the growth of roots and shoots of plant seedlings.
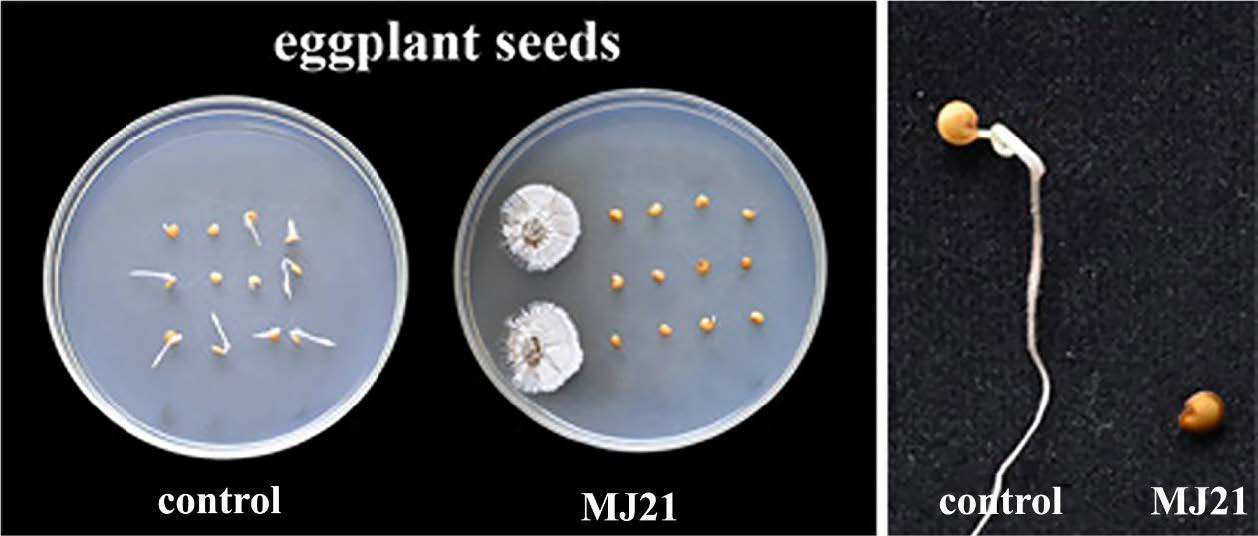
Figure 3. Appearance of plant seedlings 5 days after seeds were grown in a petri dish containing a 5-day-old culture of Streptomyces isolate MJ21 in comparison with the seeds grown on nutrient agar (NA) without bacteria.
Identification of putative pathogenic Streptomyces by PCR
The set of primers Nf and Nr was designed to amplifiy a 700-bp fragment of the nec1 intergenic region, 385 bp of the txtAB intergenic region and 392 bp of the tomA intergenic region. Isolate MJ21 produced a PCR product for two of the three pathogenic gene sets of primers tested. The Streptomyces isolate MJ21 only yielded PCR products for the tomA and txtAB set of primers, which are related to the production of a tomatinase enzyme and thaxtomin A. Isolate MJ27, MM03 and PE04 did not yield the three expected PCR gene size products (Table 2). When the sequences were deposited, the tomatinase gene clustered at 98.90-100% with the tomA gene of Streptomyces sp. strain st101 (accession number AIX97194.1) and S. scabies (accession number ACJ682220.1), and thaxomin synthetase A clustered at 96.15-100% with txtAB gene of S. scabies (accession number ACJ74085.1), S. acidiscabies (accession number AFK93859.1) and S. turgidiscabies (accession number ACJ74077.1). Furthermore, the Streptomyces isolate MJ21 has a toxigenic region that is associated with the tomA and txtAB genes (accession number OK064161 and OK064162, respectively).
Table 2. Detection of the virulence-related genes of Streptomyces from potato tubers by PCR.
|
Streptomyces isolates |
Virulence-related genes |
||
|
Nec1 gene |
txtAB gene |
tomA gene |
|
|
MJ20 |
- |
- |
- |
|
MJ21 |
- |
+ |
+ |
|
MM03 |
- |
- |
- |
|
PE4 |
- |
- |
- |
(+) presence; (-) absence
16S rRNA gene amplification
The sequence was compared with those in the GenBank (Table 3). The neighbour-joining dendrogram confirmed that MJ21 is a member of the genus Streptomyces, dendogram most closely related to S. scabies ATCC 49173 (accession number NR116531) and S. scabies RL-34 (accession number NR025865) with a sequence similarity of 99.93 – 100.00 %. Therefore, the bacterial isolate MJ21 was identified as Streptomyces scabies (accession number MW683488) (Figure 4).
Table 3. List of Sequences and GenBank accession numbers of Streptomyces species used in phylogenetic analyses.
|
Streptomyces species |
Strain |
Source |
Origin |
|
Bacterial strain |
|
|
|
|
|
MJ21 |
Solanum tuberosum |
Thailand |
|
Reference strains |
|
|
|
|
Streptomyces scabiei |
RL-34 |
Solanum tuberosum |
- |
|
Streptomyces scabiei |
ATCC 49173 |
Solanum tuberosum |
Canada |
|
Streptomyces europaeiscabiei |
KACC 20186 |
Solanum tuberosum |
- |
|
Streptomyces europaeiscabiei |
CFBP 4497 |
Solanum tuberosum |
- |
|
Streptomyces bottropensis |
ATCC 25435 |
Solanum tuberosum |
- |
|
Streptomyces stelliscabiei |
CFBP 4521 |
Solanum tuberosum |
France |
|
Streptomyces turgidiscabiei |
ATCC 700248 |
Solanum tuberosum |
- |
|
Streptomyces turgidiscabiei |
266 |
Solanum tuberosum |
- |
|
Streptomyces puniciscabiei |
S77 |
Solanum tuberosum |
Korea |
|
Streptomyces acidiscabies |
RL-110 |
Solanum tuberosum |
- |
|
Streptomyces acidiscabies |
- |
Solanum tuberosum |
Mexico |
|
Streptomyces tendae |
175 |
Brown semidesert soil |
Mongolia |
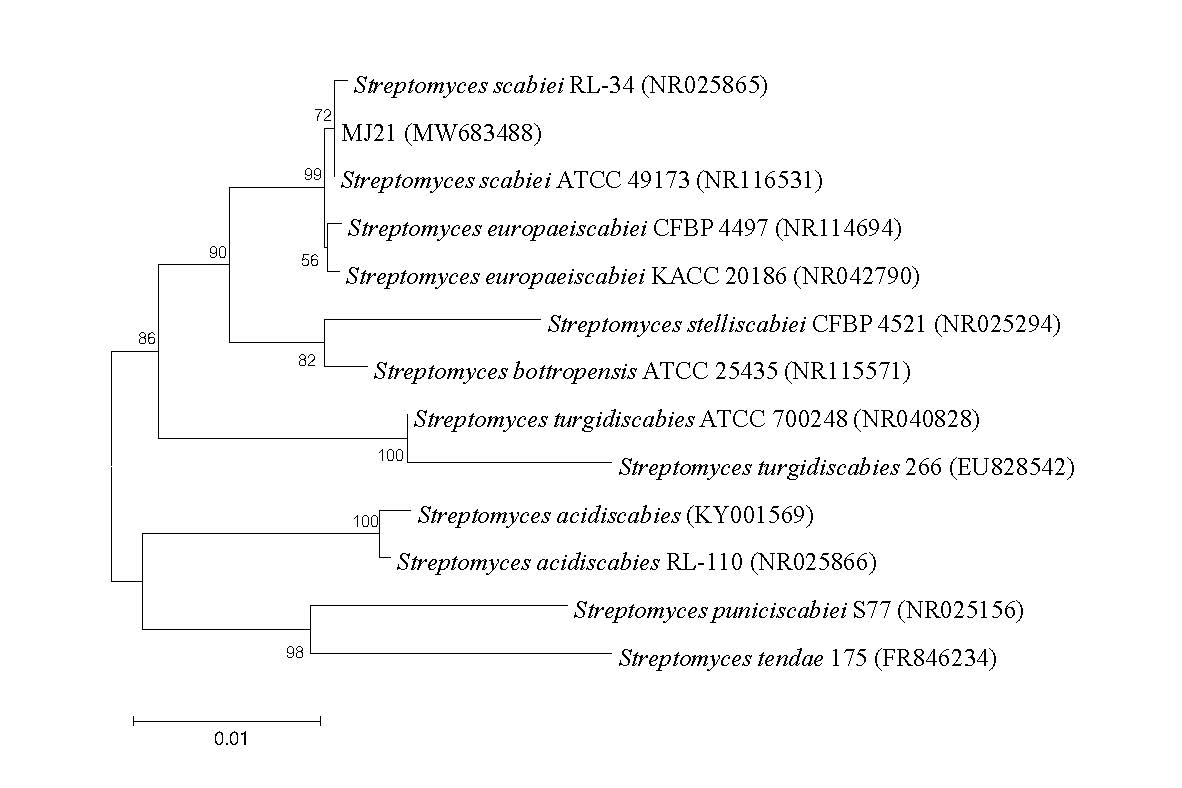
Figure 4. Phylogenetic tree of Streptomyces isolate MJ21 based on the 16S rRNA gene sequences obtained by the Neighbor-Joining method. The numbers at the branching points are the percentages of occurrence in 1000 bootstrapped trees.
DISCUSSION
Potato scab is a worldwide disease caused by different Streptomyces species. The pathogenicity test on potato revealed that Streptomyces isolate MJ21 induced damage to the general skin and caused symptoms in a larger proportion of the surface of the tubers. The current study showed that Streptomyces MJ21 causing potato scab disease can be classified as Streptomyces scabies. The S. scabies MJ21 formed flexuous spore chains, which are sympodially branched and rarely curled at the tip as similarly reported by Lambert and Loria (1989). The number of Streptomyces species is increasing as cited by literature listing more than 10 Streptomyces species which have been described as the causal agents of the potato scab complex disease in different geographical regions worldwide.
S. scabies MJ21 produced PCR products for the tomA and txtAB sets of primers, which are related to the production of a tomatinase enzyme and thaxtomin A, respectively. The characteristic PAI, also a saponinase like gene (tomA), apparently lies outside of nec1, more distant from txtAB. Nevertheless, some species typically have a different composition of PAI genes. Wanner (2007) reported that Streptomyces sp. Idaho X nearly always had tomA but not nec1. The tomA gene from the Streptomyces PAI has led to speculation as to virulence in potato diseases or its involvement in pathogenicity (Bouarab et al., 2002; Kers et al., 2005; Wanner, 2006). Flores-Gonzalez et al. (2008) and Wanner (2009) reported that different pathogenic Streptomyces species lack nec1 or tomA. Actually, most researchers suggested that nec1 and tomA genes are associated with pathogenicity but they weren’t the primary determinants. Some pathogen strains lacking one or two of these genes were reported by Wanner (2009), Dees et al. (2013) and Leminger et al. (2013). According to Karagöz and Kotan (2017), many researchers have formed a consensus that scab-causing strains most often produce thaxtomin but this may not be already operative. In Korea, Streptomyces scabiei, S. acidiscabies and S. turgidiscabies were pathogenic on progeny tuber and determined by the production of thaxtominA and homologs of nec gene (Park et al 2003). Streptomyces scabiei appeared more frequently in weakly acidic to neutral soils rather than strongly acid soil (Tashiro et al., 2012). Moreover, the existence of nec1 and tomA genes were also reported in non-pathogenic strains. On the other hand, Loria et al. (2006), Flores-Gonzalez et al. (2008) and Wanner et al. (2004) investigated some scab-causing Streptomyces species that may not have txtAB. Lapaz et al., (2017) Obtained Streptomyces isolates from potato-producing regions in Uruguay that found Streptomyces scabiei, S. acidiscabies, and S. europaeiscabiei. These isolates carried the txtA, txtB, tomA and nec1 genes, commonly associated with pathogenicity in Streptomyces and characteristic of PAI. However, as previously reported by Gartemann et al. (2003) potato also produces saponins, which could be the substrate for the putative tomatinase in the PAI. Further characterization of the identified Streptomyces strain and the phytotoxic substance of plants produced is currently progressing to better understand how the substance can cause disease. A particularly interesting finding of this study was that Streptomyces scabies MJ21 displayed a severe pathogenic phenotype against different plant hosts exhibited as inhibition of plant seed germination presumably by production of a novel secreted phytotoxic substance. As previously reported by Cao et al. (2012), a Streptomyces strain causing deeply-pitted lesions on potato tubers produced an 18-membered macrolide called borelidin, inhibited the growth of shoots and roots of radish seedlings and induced necrosis of potato tuber slices. The tomA and nec1 genes are also present in a wide range of potato common scab - inducing Streptomyces strains (Seipke and Loria, 2008).
CONCLUSION
Streptomyces isolate MJ21 causing potato scab disease is Streptomyces scabies. This is identifying and detecting a virulence gene of S. scabies in Chiang Mai, Thailand. The current research extends the knowledge of the presence of this microorganism in this area. Thorough characterization of bacterial pathogens of the potato common scab will deepen our cognition of the pathogenicity and this has important implications for the development of long-term control strategies that can be effective for the management of disease in potato growing regions.
ACKNOWLEDGEMENTS
This research was partially supported by the Innovation Agriculture Research Center, Faculty of Agriculture, Chiang Mai University and the Center of Excellence on Agricultural Biotechnology: (AG-BIO/MHESI), Bangkok, Thailand.
AUTHOR CONTRIBUTIONS
Conceptualization: Angsana Akarapisan, Methodology: Athidtaya Kumvinit, Supaporn Falert, Writing-review and editing: Angsana Akarapisan, Athidtaya Kumvinit, Supaporn Falert, Wichai Kositratana, All authors have read and agreed to the published version of the manuscript.
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
REFERENCES
Alejo, A., Burgueño, E., Maldonado, L.A., Herrera, G., Felix, R., and Quintana, E.T. 2019. In vitro effect of the crude extract of a potato common scab streptomycete in Sinaloa, Mexico. Revista Argentina de Microbiología. 51: 363-370.
Atiq, M., Khalid, A.R., Hussian, W., Nawaz, A., Asad, S., and Ahmad, T.M. 2013. Genetic potential of potato germplasm against common scab disease caused by Streptomyces scabies. Pakistan Journal of Phytopathology. 25: 27–30.
Bouarab, K., Melton, R., Peart, J., Baulcombe, D., and Osbourn, A. 2002. A saponin-detoxifying enzyme mediates suppression of plant defences. Nature. 418: 889-892.
Bouchek-Mechiche, K., Gardan, L., Normand, P., and Jouan, B. 2000. DNA relatedness among strains of Streptomyces pathogenic to potato in France: Description of three new species S. europaeiscabiei sp. nov. and S. stelliscabiei sp. nov. associated with common scab and S. reticuliscabiei sp. nov. associated with netted scab. International Journal of Systematic and Evolutionary Microbiology. 50: 91-99.
Cao, Z., Khodakaramian, G., Arakawa, K., and Kinashi, H. 2012. Isolation of borrelidin as a phytotoxic compound from a potato pathogenic Streptomyces strain. Bioscience Biotechnology and Biochemistry. 76: 353-357.
Cheng, H.R., and Jiang, N. 2006. Extremely rapid extraction of DNA from bacteria and yeasts. Biotechnology Letters. 28: 55-59.
Dees, M.W., Sletten, A., and Hermansen, A. 2013. Isolation and characterization of Streptomyces species from potato common scab lesions in Norway. Plant Pathology. 62: 217-225.
Flores-González, R., Velasco, I., and Montes, F. 2008. Detection and characterization of Streptomyces causing potato common scab in Western Europe. Plant Pathology. 57: 162-169.
Fyans, J.K., Bown, L., and Bignell, D.R.D. 2016. Isolation and characterization of plant- pathogenic Streptomyces species associated with common scab-infected potato tubers in Newfoundland. Phytopathology. 106: 123-131.
Gartemann, K.H., Kirchner, O., Engemann, J., Grafen, I., Eichenlaub, R., and Burger, A. 2003. Clavibacter michiganensis subsp. michiganensis: First steps in the understanding of virulence of a Gram-positive phytopathogenic bacterium. Journal of Biotechnology. 106: 179-191.
Ismail, S., Jiang, B., Nasimi, Z., Inam-ul-Haq, M., Yamamoto, N., Ofori, D.A., Khan, N., Arshad, M., Abbas, K., and Zheng, A. 2020. Investigation of Streptomyces scabies causing potato scab by various detection techniques, its pathogenicity and determination of host-disease resistance in potato germplasm. Pathogens. 760: 1-26.
Karagöz, K. and Kotan, R. 2017. Identification and characterization of some Streptomyces species isolated from symtomatic potatoes in Erzurum province of Turkey. Eastern Anatolian Journal of Science. 3: 27-37.
Kers, J.A., Cameron, K.D., Joshi, M.V., Bukhalid, R.A., Morello, J.E., Wach, M.J., Gibson, D.M., and Loria, R. 2005. A large, mobile pathogenicity island confers plant pathogenicity on Streptomyces species. Molecular Microbiology. 55: 25-1033.
Kittipadakul, P., Jaipeng, B., Slater, A., Stevenson, W., and Jansky, S. 2016. Potato production in Thailand. American Journal of Potato Research. 93: 380-385.
Lambert, D.H. and Loria, R. 1989. Streptomyces scabies sp. nov., nom. rev. International Journal of Systematic and Evolutionary Microbiology. 39: 387-392.
Lapaz, M.I., Huguet, J.C., Siri, M.I., Verdier, E., Loria, R., and Pianzzola, M.J. 2017. Genotypic and phenotypic characterization of Streptomyces species causing potato common scab in Uruguay. Plant Disease. 101: 1362-1372.
Lehtonen, M.J., Rantala, H., Kreuze, J.F., Bång, H., Kuisma, L., Koski, P., Virtanen, E., Vihlman, K., and Valkonen, J.P.T. 2004. Occurrence and survival of potato scab pathogens (Streptomyces species) on tuber lesions: quick diagnosis based on a PCR-based assay. Plant Pathology. 53: 280-287.
Leiminger, J., Frank, M., Wenk, C., Poshenrieder, G., Kellermann, A., and Schwarzfisher, A. 2013. Distribution and characterization of Streptomyces species causing potato common scab in Germany. Plant Pathology. 62: 611-623.
Lerat, S., Simao-Beaunoir, A.M., and Beaulieu, C. 2009. Genetic and physiological determinants of Streptomyces scabies pathogenicity. Molecular Plant Pathology. 10: 579-585.
Loria, R., Kers, J., and Joshi, M. 2006. Evolution of plant pathogenicity in Streptomyces. Annual Review of Phytopathology. 44: 469-487.
Mingma, R., Pathom-aree, W., Trakulnaleamsai, S., Thamchaipenet, A., and Duangmal, K. 2014. Isolation of rhizosperic and roots endophytic actinomycetes from Leguminosea plant and their acitivities to inhibit soybean pathogen, Xanthomonas campestris pv. glycine. World Journal of Microbiology and Biotechnology. 30: 271-280.
Miyajima, K., Tanaka, F., Takeuchi, T., and Kuninaga, S. 1998. Streptomyces turgidiscabies sp. nov. International Journal of Systematic and Evolutionary Microbiology. 48: 495-502.
Park, D.H., Yu, Y.M., Kim, J.S., Cho, J.M., Hur, J.H., and Lim, C.K. 2003. Characterization of Streptomycetes causing potato common scab in Korea. Plant Disease 87:1290-1296.
Seipke, R.F. and Loria, R. 2008. Streptomyces scabies 87-22 possesses a functional tomatinase. Journal of Bacteriology. 190: 7684-7692.
Shutsrirung, A., Chromkaew, Y., Pathom-aree, W., Choonluchanon, S. and Boonkerd, N. 2013. Diversity of endophytic actinomycetes in mandarin grown in northern Thailand, their phytohormone production potential and plant growth promoting activity. Soil Science and Plant Nutrition. 59: 322-330.
Song, J., Lee, S.C., Kang, J.W., Baek, H.J., and Suh, J.W. 2004. Phylogenetic analysis of Streptomyces spp. isolated from potato scab lesions in Korea on the basis of 16S rRNA gene and 16S-23S rDNA internally transcribed spacer sequences. International Journal of Systematic and Evolutionary Microbiology. 54: 203-209.
Song, Z., Liu, K.Q., Lu, Q.X., Yu, J., Ju, R.C., and Liu, X.L. 2011.. Isolation and characterization of a potential biocontrol Brevibacillus laterosporus. African Journal of Microbiology Research. 5: 2675-2681.
Soe, K.M., Bhromsiri, A., and Karladee, D. 2010. Effects of selected endophytic actinomycetes (Streptomyces sp,) and Bradyrhizobia from Myanmar on growth, nodulation, nitrogen fixation and yield of different soybean varieties. Chiang Mai University Journal of Natural Sciences 9: 95–109.
St-Onge, R., Goyer, C., Con, R., and Filion, M. 2008. Genetic diversity of Streptomyces spp. causing common scab of potato in eastern Canada. Systematic and Applied Microbiology. 31:474–484.
Takeuchi, T., Sawada, H., Tanaka, F., and Matsuda, I. 1996. Phylogenetic Analysis of Streptomyces spp. causing potato scab based on 16S rRNA sequences. International Journal of Systematic and Evolutionary Microbiology. 46: 476-479.
Tashiro, N., Manabe, K., Saito, A., and Miyashita, K. 2012. Identification of potato scab-causing Streptomyces sp. occurring in strongly acidic soils in Saga Prefecture in Japan. Journal of General Plant Pathology. 78: 353-359.
Wanner, L.A. 2004. Field isolates of Streptomyces differ in pathogenicity and virulence on radish. Plant Disease. 88: 785-796.
Wanner, L.A. 2006. A survey of genetic variation in Streptomyces isolates causing potato common scab in the United States. Phytopathology. 96: 1363-1371.
Wei, Q., Li, J., Yang, S., Wang, W., Min, F., Guo, M., Zhang, S., Dong, X., Hu, L., Li, Z., and Wang, Z. 2020. Streptomyces rhizophilus causes potato common scab disease. Plant Disease. 106: 266-274.
Wanner, L.A. 2007. A new strain of Streptomyces causing common scab in potato. Plant Disease. 91: 352-359.
Wanner, L.A. 2009. A patchwork of Streptomyces species isolated from potato common scab lesions in North America. American Journal of Potato Research. 86: 247-264.
Wanner, L.A., and Kirk, W.W. 2015. Streptomyces – from basic microbiology to role as a plant pathogen. American Journal of Potato Research. 92: 236-242.
Yang, M., Wang, R., Du, W., Gong, C., Yu, D., Zhao, W., Song, S., and Zhang, H. 2018. The pathogenic Streptomyces species causing potato scab in Yunnan Province. Acta Phytopathologica Sinica. 48: 445–454.
OPEN access freely available online
Natural and Life Sciences Communications
Chiang Mai University, Thailand.
Angsana Akarapisan1, 2, *, Athidtaya Kumvinit1, 2, Supaporn Falert1, 2, and Wichai Kositratana2, 3
1 Division of Plant Pathology, Department of Entomology and Plant Pathology, Faculty of Agriculture, Chiang Mai University, 50200 Chiang Mai, Thailand.
2 Center of Excellence on Agricultural Biotechnology: (AG-BIO/MHESI), Bangkok 10900, Thailand.
3 Center for Agricultural Biotechnology, Kasetsart University, Kamphaeng Saen Campus, Nakhon Pathom 73140, Thailand.
Corresponding author: Angsana Akarapisan, E-mail: angsana.aka@gmail.com
Total Article Views
Editor: Pachara Sattayawat,
Chiang Mai University, Thailand
Article history:
Received: August 10, 2022;
Revised: February 5, 2023;
Accepted: February 7, 2023;
Published online: February 20, 2023

