
Immune Response to Common Bacteria Causing Sepsis in Myanmar Workers in Northeast Thailand: A Preliminary Study
Myint Myint Sein, Arnone Nithichanon, Narisara Chantratita, Chidchamai Kewcharoenwong* and Ganjana Lertmemongkolchai*Published Date : April 3, 2023
DOI : https://doi.org/10.12982/NLSC.2023.026
Journal Issues : Number 2, April-June 2023
ABSTRACT
Migrant workers from Myanmar living in Thailand have significantly increased, these migrant workers may display health risk of common bacterial infections and host immune response of Myanmar workers to those common bacterial infections is still not well known. In this study, we observed antibodies and interferon-gamma to the common bacteria causing sepsis in this area, such as Escherichia coli, Klebsiella pneumonia and Pseudomonas aeruginosa including Burkholderia pseudomallei which is pathogenic and endemic in this area. The results showed that Myanmar workers decreased cellular function of interferon-gamma response, compared with native Thai workers. But the humoral response of human IgG showed no significant difference between Myanmar and Thai workers. Even though E. coli and K. pneumonia induced lower level of plasma IgG response than the other two bacteria, but host plasma could effectively kill and clear the bacteria within an hour in vitro. Interestingly, the level of anti-Hcp 1, which is a diagnostic marker for B. pseudomallei, was low in Myanmar workers suggesting no exposure to this organism. However, the sample size should be increased to reach the conclusion. Our study could provide useful information for Myanmar migrant workers to promote safe working and awareness of health risks to bacterial infections.
Keywords: Burkholderia pseudomallei, Hcp-1, Interferon gamma, Plasma IgG, Myanmar migrant workers
Funding: The authors acknowledged the funding provided by Faculty of Associated Medical Sciences, Khon Kaen University, Khon Kaen, Thailand.
Citation: Sein, M.M., Nithichanon, A., Chantratita, N., Kewcharoenwong, C., and Lertmemongkolchai, G. 2023. Immune response to common bacteria causing sepsis in Myanmar workers in Northeast Thailand: A preliminary study. Nat. Life Sci. Commun. 22(2): e2023026.
INTRODUCTION
Bacterial infection is one of the common causes of global deaths. In high income countries, Escherichia coli is the most infected bacteria followed by Staphylococcus aureus, Streptococcus species, Klebsiella pneumonia, Pseudomonas species and other gram-negative bacteria in blood stream infection (Cukic et al. 2016, Laupland et al. 2016, Kern et al. 2020). In Southeast Asia Region, especially low-income countries, bacterial infections like tuberculosis, respiratory infections and diarrheal diseases are important cause of death. Now-a-day, Thailand hosts many migrant workers from neighboring countries including Myanmar. These Myanmar workers may have consequence of infectious diseases (WHO 2008, Coker et al. 2011, Aung et al. 2020). According to the previous studies in Myanmar during 2015-2016, blood stream infection was mainly caused by Salmonella enterica, E. coli, K. pneumonia, S. aureus and followed by yeasts, B. pseudomallei and S. agalactiae (Myat et al. 2014, Myat et al. 2020). In a previous study in Northeast Thailand, top five common bacteria of pneumonia, urinary tract infection and blood stream infections were Acinetobacter baumannii, E. coli, K. pneumoniae, P. aeruginosa, and S. aureus (Phodha et al. 2019).
Even though bacterial infections can be treatable by antibiotics, now-a-days it becomes global health problem with high mortality due to increasing resistance to current drugs. Therefore, host immune system becomes more priority to prevent hosts from infections. Human immune response to infection can vary according to environmental factors, race, and ethnicity (Piruzian et al. 2009, Liston et al. 2016, Brodin et al. 2017, Nahid et al. 2018). The host immune response is composed of rapid response of innate and specific response of adaptive immune system. Cellular effector molecule of interferon-gamma (IFN-γ) enhances innate immune cells for phagocytosis and enhances adaptive immune cells for antibody production (Kak et al. 2018). Effector antibody of IgG to specific pathogen plays a major role in pathogen clearance (Shtrichman et al. 2001, Abbas et al. 2018).
In this study, we aimed to test human background immunity to common bacteria by determining IFN-γ and IgG response of Myanmar workers living in Thailand and compared with Thai workers. This study may provide the basic immune status of Myanmar workers to common bacteria in working area that can be useful for the workers who tend to work in Thailand in knowing health risks of working environment and taking awareness or prevention.
MATERIALS AND METHODS
Common bacterial sepsis report
Information of patients with bacterial culture and identification requested during 2018 to 2019 were obtained from Microbiology Unit, Department of Medical Technology and Pathology, Nakhon Phanom Hospital with human research ethic approval number NP-EC11-No.1/2559. Only cases with bacterial culture positive were documented. Repeated samples from the same case with the same organism were excluded. Coagulase-Negative Staphylococci was determined as true pathogen only in case of hemoculture.
Human sample collection
Heparinized blood samples from Myanmar and Thai healthy donors were collected with written informed consent. This study was approved by Khon Kaen University Ethics Committee for Human Research (approval number HE622097). In the study, post hoc analysis was used to determine effective sample size and detection of differences between two groups of people, sample size of at least 21 participants per group was necessary (α = 0.05, β = 0.20). IFN-γ to K. pneumoniae in which to total number of sample 12 (6/group) was necessary to detect the differences. However, due to the limitation of time and specimen collection, this sample size was informed by the estimated effect size of the intervention on other parameter, such as the power for detection of differences between two groups of IFN-γ to E. coli and P. aeruginosa, by post hoc analysis, showed 44.7% and 71.5% respectively. So, further study with increasing sample size of these two parameters is still needed.
Whole blood culture was performed immediately after blood collection, the remaining plasma samples were stored at -80°C until use. Leftover plasma samples from septicemic melioidosis (Bp sepsis) cases and melioidosis recovered (Bp recovery) cases were obtained from the previous study (Nithichanon et al. 2018).
Strain of bacteria and preparation
E. coli ATCC25922, K. pneumonia ATCC700603, P. aueroginosa ATCC27853 and B. pseudomallei K96243 were used in this study and prepared by the previously reported protocol (Piasecka et al. 2018).
Whole blood culture and detection of human IFN-γ
Heparinized whole blood from healthy donors (number of lymphocytes and monocytes were adjusted to 1.8 × 106/ml) were cultured at 37°C in 5% CO2 with cell culture medium control (RPMI1640 medium supplemented with 10% fetal bovine serum, FBS), 3 µg/ml phytohemagglutinin (Aung et al. 2020), 107 CFUs paraformaldehyde (PFA)-killed E. coli, K. pneumoniae, P. aeuroginosa, or B. pseudomallei. Supernatants were collected at 48 hours after incubation and stored at -80°C. Level of producing human IFN-γ in the culture supernatant were quantified by using a human IFN-γ ELISA kit (BD Biosciences, USA), following the manufacturer’s instructions.
Detection of human plasma IgG antibody by indirect ELISA
Human plasma IgG has been detected by previously published (Capelli et al. 2018). PFA-fixed intact bacteria at 107 CFUs were coated onto 96-well polystyrene plate (Nunc, Denmark) with carbonate bicarbonate buffer pH 9.6 overnight at 40 °C. The plates were washed with washing buffer, 0.1% Tween-20 in PBS pH 7.4, before blocked with 10% FBS in PBS at room temperature for 2 hours. Heparinized plasma samples were diluted at 1:300 with assay diluent (0.05% Tween-20, 10% FBS in PBS). The solution was removed from the pre-coated plates before adding diluted plasma samples and incubated at room temperature for 2 hours. After five times washing, a detection antibody mixture (1:10,000 biotinylated goat anti-human IgG (BD Biosciences, USA), 1:1,000 HRP conjugated streptavidin (BD Biosciences, USA) diluted in assay diluent) was added and incubated at room temperature for 1 hr. After seven times washing, 3,3',5,5'-tetramethylbenzidine (TMB) substrate (BD Biosciences, USA) was added and incubated at room temperature for 10 mins, before stopping the reaction with 2 N H2SO4. Absorbance of each well was read at 450 nm by ELISA reader. Results were shown as absorbance index and calculated as the formula below (Gourlay et al. 2013, Nithichanon et al. 2015).
Absorbance Index=(absorbance test-absorbance uncoat)
(absorbance uncoat)
Bacterial plasma killing assay
The protocol was adopted from the previous publication (van der Maten et al. 2017). Bacteria were grown to mid-log phase at 37°C in LB broth and adjust by MacFarland unit 0.3 equivalents to 108 CFU/ml of bacteria. Plasma samples from 2 Thai healthy adult volunteers were prepared as normal plasma and heat-inactivated plasma at 56°C for an hour.
Live bacteria of 1,000 CFUs were culture with FBS, normal human plasma, or heat-inactivated human plasma at 37°C for 1, 2 and 4 hours. Bacteria number enumeration was performed at baseline number and at indicated hour of incubation by standard plating technique on LB agar. Data was analyzed and shown as % bacterial survival, a percentage of bacterial number at indicated time point per baseline number.
Statistical analysis
Statistical analysis was carried out using GraphPad Prism version 6 (GraphPad, USA). Statistical differences were determined using one-way ANOVA, two-way ANOVA, and Mann-Whitney test for independent groups of non-gaussian distribution data. Comparison of Data correlation was analyzed using the Pearson correlation test. Statistically significant differences were considered at P < 0.05.
RESULTS
Top 5 common bacterial organisms were reported from Nakhon Phanom Hospital during 2018 and 2019.
Information of patients with bacterial culture and identification requests during 2018 to 2019 was obtained from Microbiology Unit, Department of Medical Technology and Pathology, Nakhon Phanom Hospital. Total case number with bacterial culture positive in 2018 was 7,047 cases, in 2019 was 6,903 cases. The top 5 bacteria isolated from each type of clinical specimens during 2018 to 2019 are shown in Table 1.
Table 1. Top 5 bacteria isolated from different sources of infection from patients at Nakhon Phanom Hospital who admitted during 2018 to 2019.
|
Specimen types |
2018 |
2019 |
|||
|
Bacterial identification |
Reported cases (%) |
Bacterial identification |
Reported cases (%) |
||
|
Hemoculture |
Escherichia coli Burkhoderia pseudomallei Coagulase Negative Staphylococcus Klebsiella pneumoniae Staphylococcus aureus |
378 (20.1%) 171 (9.1%) 132 (7.0%) 131 (7.0%) 114 (6.1%) |
Escherichia coli Coagulase Negative Staphylococcus Burkhoderia pseudomallei Klebsiella pneumoniae Staphylococcus aureus |
512 (21.7%) 338 (14.3%) 182 (7.7%) 166 (7.0%) 166 (7.0%) |
|
|
Sputum
|
Klebsiella pneumoniae Acinetobacter baumannii Pseudomonas aeruginosa Staphylococcus aureus Streptococcus pneumoniae |
400 (20.4%) 234 (12.0%) 209 (10.7%) 188 (9.6%) 144 (7.4%) |
Klebsiella pneumoniae Staphylococcus aureus Pseudomonas aeruginosa Acinetobacter baumannii Streptococcus pneumoniae |
389 (22.5%) 230 (13.3%) 226 (13.1%) 195 (11.3%) 108 (6.3%) |
|
|
Local wound and pus |
Staphylococcus aureus Escherichia coli Klebsiella pneumoniae Pseudomonas aeruginosa Enterobacter species |
314 (19.6 %) 189 (11.8%) 129 (8.0%) 91 (5.7%) 64 (4.0%) |
Staphylococcus aureus Escherichia coli Pseudomonas aeruginosa Klebsiella pneumoniae Burkhoderia pseudomallei |
331 (23.1%) 185 (12.9%) 125(8.7%) 118 (8.2%) 84 (5.9%) |
|
|
Urine, Catheter |
Escherichia coli Enterococcus faecalis Klebsiella pneumoniae Enterococcus faecium Streptococci Group D, Non-Enterococci |
545 (34.0%) 324 (20.2%) 149 (9.3%) 100 (6.2%) 44 (2.7%) |
Escherichia coli Enterococcus faecalis Klebsiella pneumoniae Enterococcus faecium Streptococci Group D |
460 (33.2%) 328(23.6%) 133 (9.6%) 122 (8.8%) 47 (3.4%) |
|
E. coli was the most common isolated bacteria from hemoculture with a very dominate proportion compared to other bacteria which were mostly less than 10%. B. pseudomallei, a deadly bacterium responsible for causing melioidosis in humans, was reported as the second and the third most bacteria found from hemoculture in 2018 and 2019, respectively. K. pneumoniae was dominated in terms of bacteria found in sputum specimens and the following bacterial species were A. baumannii, P. aeuroginosa. In case of skin infection (local wound and pus) specimens, S. aureus was dominated following by E. coli, K. pneumonia, P. aeuroginosa and Enterobacter species or B. pseudomallei while urine or catheter specimens were mainly reported positive for E. coli and Enterococcus faecalis.
Therefore, E. coli, K. pneumoniae, P. aeuroginosa and S. aureus are bacteria which can be found as pathogens in clinical specimens. Whereas the occurrence of B. pseudomallei infection needs to be monitored due to its highly pathogenic.
Thai healthy workers developed more potent IFN-γ response against the common infectious bacteria than Myanmar workers, with the same level of plasma IgG.
Healthy workers with no sign of infection in the previous 3 months were recruited into this study to compare in vitro immune responses against common infectious bacteria between Myanmar and Thai workers. Heparinized whole blood samples were collected from 20 Myanmar and 8 Thai healthy individuals. Demographic data was shown in Table 2, briefly, there were no significant differences in age, sex, total white blood cell count, neutrophil or lymphocyte, but monocyte number was significantly increased in Myanmar individuals.
Table 2. Subject characteristics and demographics.
|
Parameters |
Myanmar (n = 20) |
Thai (n = 8) |
P-value |
|
Age, yrs. (median, range) Sex (n, % female) Occupation (n, %)
Duration of stay in Thailand, yrs. (Median, range) White blood cell count (103 cells/µL) (median, range) Neutrophil Lymphocyte Monocyte |
36 (26 – 51) 10 (50%)
3 (15) 14 (70) 3 (15) 8.5 (4-24)
6.6 (3.7 – 9.0)
3.21 (1.39-5.73) 2.6 (1.58-4.12) 0.45 (0.26-0.62) |
43 (29 – 47) 7 (87%)
- - - -
5.9 (3.9 – 8.2)
3.5 (1.88-4.34) 2.02 (1.12-3.0) 0.35 (0.19-0.4) |
0.6435 0.0987
NA NA NA NA
0.1471
0.4228 0.1322 0.0066 |
To investigate the differences of immune response against common bacteria in the area including E. coli, K. pneumoniae, P. aeuroginosa, and B. pseudomallei, whole blood samples were stimulated with killed dead bacteria or phytohemagglutinin (Aung et al. 2020) as positive control, or medium control. We quantified for IFN-γ, a key proinflammatory cytokine in defense against bacterial infections, by ELISA. The results of whole blood culture from Thai and Myanmar workers revealed no background IFN-γ in cell culture medium control with the non-significant difference of response against a positive control, PHA (Figure 1A). In part of IFN-γ producing upon stimulated with dead whole bacterial cells, the level of IFN-γ from Thai worker group was significantly higher than Myanmar worker group in response to E. coli, K. pneumoniae, or P. aeruginosa (P-value < 0.01, < 0.001, and < 0.01 respectively), whereas the response of Thai worker group against B. pseudomallei tended to be higher than Myanmar worker group but did not reach statistical significance (P-value = 0.1225) (Figure 1B). These data suggest that Thai workers have developed more potent IFN-γ associated immune response against the whole cells of such common bacteria than Myanmar workers who had been worked and stayed in Thailand for years.
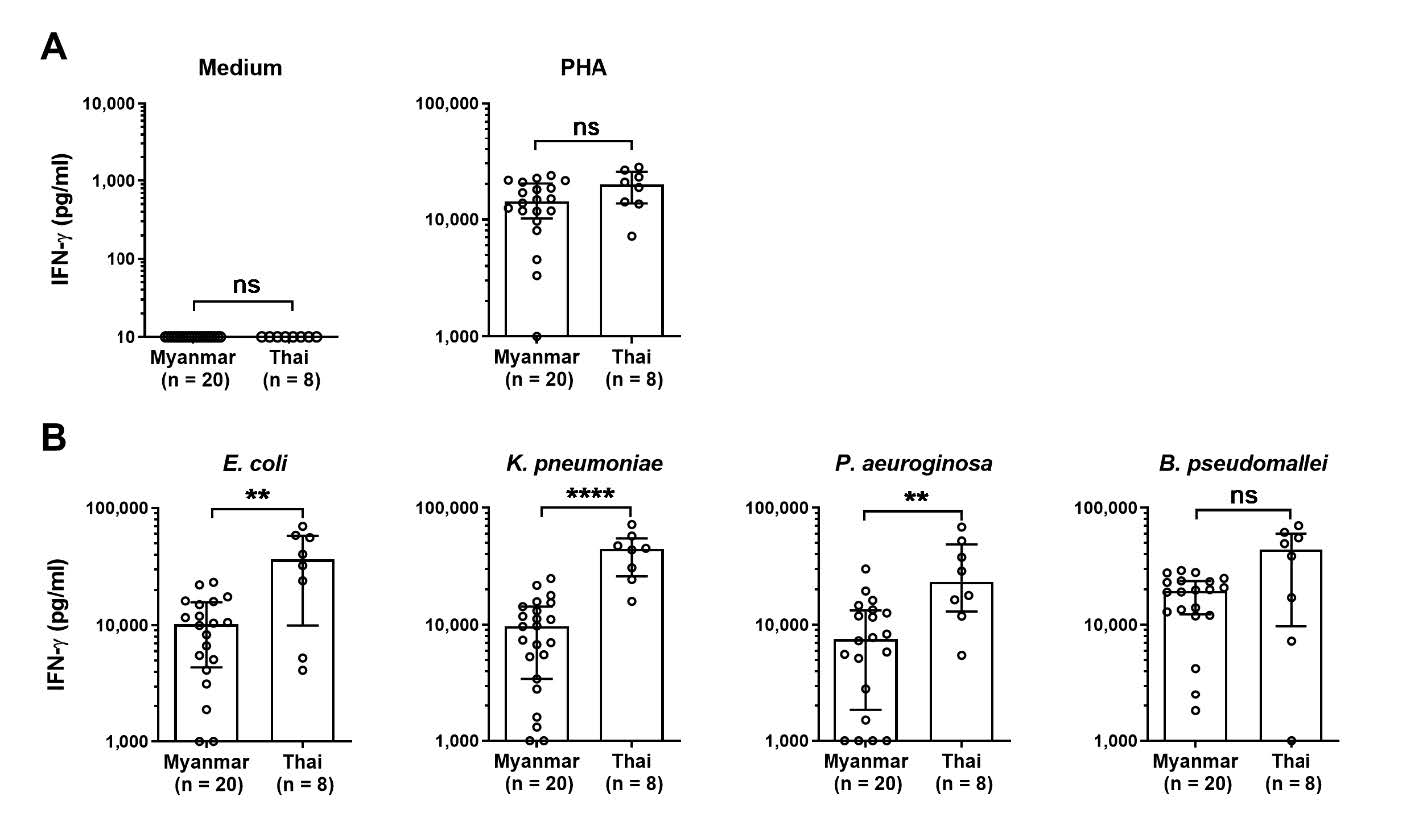
Figure 1. Comparison of IFN-γ production from Myanmar versus Thai whole blood samples upon stimulation. Heparinized whole blood samples from Myanmar (n = 20) or Thai (n = 8) healthy participants were incubated with cell culture medium, phytohemagglutinin (Aung et al. 2020), 2 x 107 CFUs of paraformaldehyde fixed E. coli, K. pneumoniae, P. aeuroginosa, or B. pseudomallei. Supernatants were collected at 48 hrs after incubation for quantification of IFN-γ by ELISA. Levels of IFN-γ after incubation are shown as scatter dot plot of each participant with bar at median and error bar at interquartile range. Statistical differences were analyzed by using Mann-Whitney test, ns; non-significant, **; P < 0.01, ****; P < 0.0001.
We then investigated whether there was any difference in plasma IgG antibody level against the bacteria, quantified by indirect ELISA and compared between Myanmar and Thai workers. The results showed that no difference of plasma IgG antibody level was found from Myanmar and Thai healthy workers against E. coli, K. pneumoniae, P. aeruginosa, or B. pseudomallei (P-value = 0.7003, 0.1384, 0.8360 and 0.6546 respectively) (Figure 2A – 2D). Since B. pseudomallei is a cause of melioidosis, a lethal infectious disease commonly found in the area (Limmathurotsakul et al. 2016, Chewapreecha et al. 2017). We addressed whether plasma IgG antibody found from Myanmar workers could be increased in relation with time of stay in Thailand. The results showed that plasma IgG antibody level against whole cell of B. pseudomallei of Myanmar workers who had stayed in Thailand for 1 – 10 years or more than 10 years, was not statistically different with P-value at 0.2939 (Figure 2E), and no significant correlation between log transformed plasma IgG level against B. pseudomallei and duration of stay in Thailand (Figure 2F).
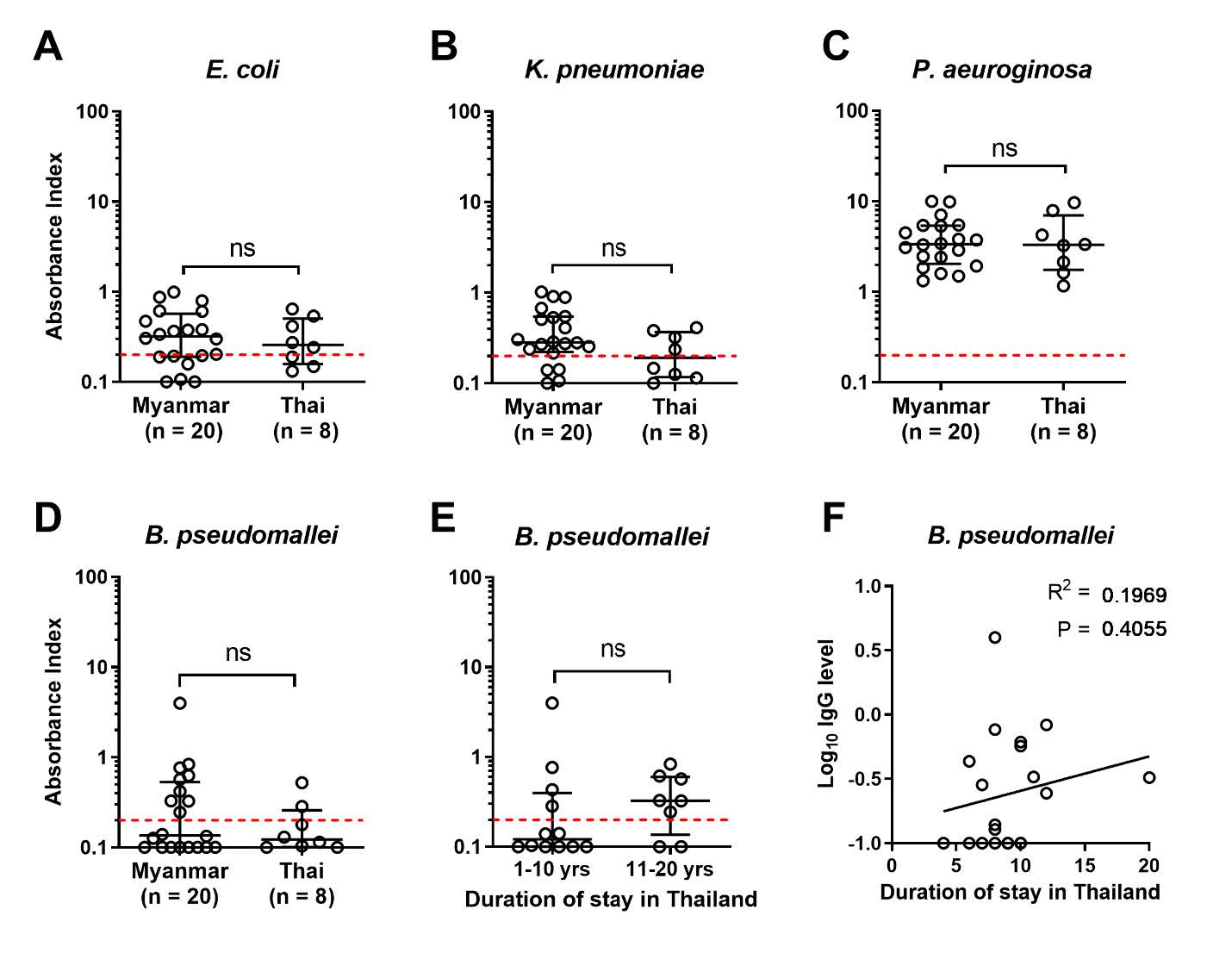
Figure 2. Comparison of Myanmar and Thai plasma IgG antibody against whole bacterial cell. Heparinized plasma samples from Myanmar (n = 20) or Thai (n = 8) healthy participants were quantified for IgG antibody level against paraformaldehyde fixed whole bacteria by ELISA. Data is shown as absorbance index of individual by dot plot with line at median and interquartile range. Plasma IgG against E. coli (A), K. pneumoniae (B), P. aeuroginosa (C), or B. pseudomallei (D) compared between Myanmar versus Thai samples. Plasma IgG level against B. pseudomallei from Myanmar samples were compared between participants who stayed in Thailand for 1 – 10 years versus 11 – 20 years. (E) Statistical differences were analyzed by using Mann-Whitney test, ns; non-significant. Correlation between IgG and duration of staying in Thailand of Myanmar participants was analyzed by Pearson correlation test (F). Red dash lines represent undetectable plasma IgG cut-off assigned by the absorbance index of average of uncoated wells plus standard deviation (average + S.D.).
To confirm that plasma specific IgG level against B. pseudomallei of Myanmar and Thai workers were not different. We tested plasma samples with Hcp-1 protein, a published diagnostic marker for melioidosis by ELISA (Burtnick et al. 2011). Plasma samples from melioidosis acute infection and recovered donors were used as positive control. The results showed similar data as using whole dead bacteria of B. pseudomallei. Of note, plasma IgG of Myanmar workers was slightly lower than Thai worker even it was not significantly different while significantly higher plasma anti-Hcp-1 IgG level found in acute melioidosis infected patients , compared to Myanmar (P-value < 0.001) or Thai healthy individuals (P-value < 0.01) (Figure 3).
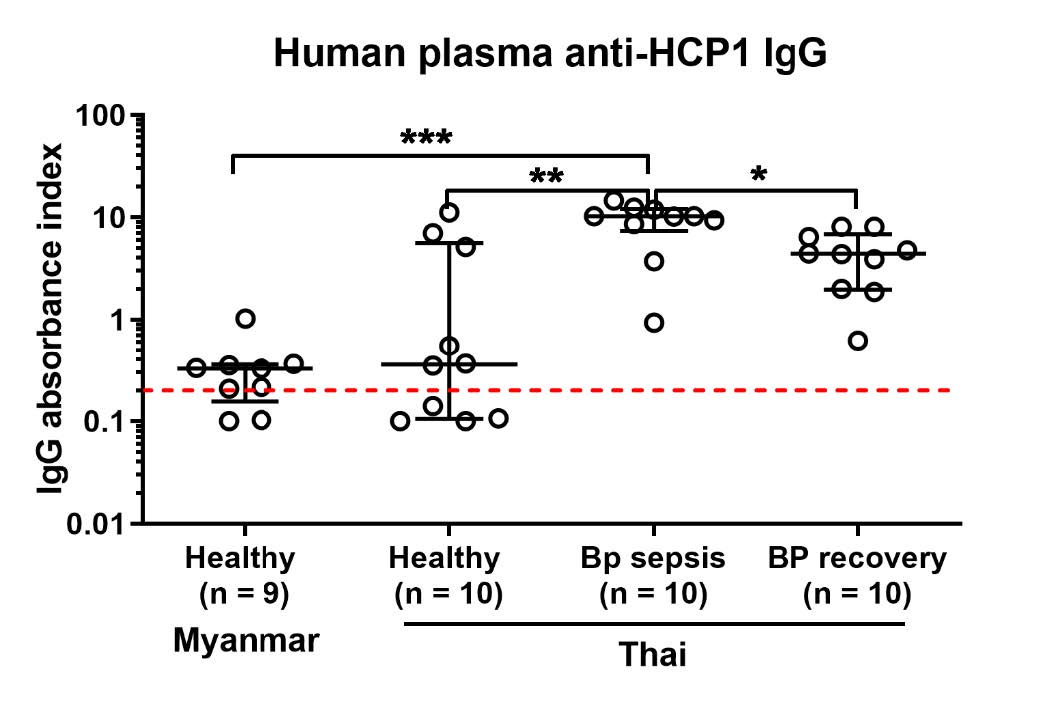
Figure 3. Comparison of Myanmar and Thai plasma IgG antibody against melioidosis diagnostic marker, Hcp-1. Heparinized plasma samples from Myanmar healthy participants (n = 10) or Thai healthy participants (n = 10), melioidosis sepsis (BP sepsis, n = 10) or melioidosis recovered participants (BP recovery, n = 10) were quantified for anti-HCP-1 IgG antibody by ELISA. Data was shown as IgG absorbance index from individual by dot plot with line at median and interquartile range. Statistical differences were analyzed by using Mann-Whitney test, ns; non-significant, *; P < 0.05, **; P < 0.01, ***; P < 0.001. Red dash lines represent undetectable plasma IgG cut-off assigned by the absorbance index of average of uncoated wells plus standard deviation (average + S.D.).
Plasma sample from healthy donors could effectively kill the bacteria via complement dependent pathway.
Generally, plasma antibodies, IgG and IgM, have a direct role on microbe killing by activation of classical pathway of complement system (Strasser et al. 2019). We investigated whether these antibodies could activate any efficiently bactericidal capacity. Two plasma samples with IgG absorbance index level against E. coli (Donor 1 = 0.241, Donor 2 = 0.413) and K. pneumoniae (Donor 1 = 0.064, Donors 2 = 0.413) were selected for bacterial plasma killing assay. Both plasma samples were aliquoted and prepared as normal plasma and heat inactivated plasma (Motley et al. 2020). Bacterial growth in heat inactivated fetal bovine serum (FBS) was used as a control to calculate the rate of bacteria proliferation in the presence of plasma protein. The results showed that normal plasma could significantly kill E. coli within 2 hours while K. pneumoniae was significantly killed within 1 hour, compared to heat inactivated FBS or plasma (Figure 4) suggesting that these antibodies contained bactericidal activities.
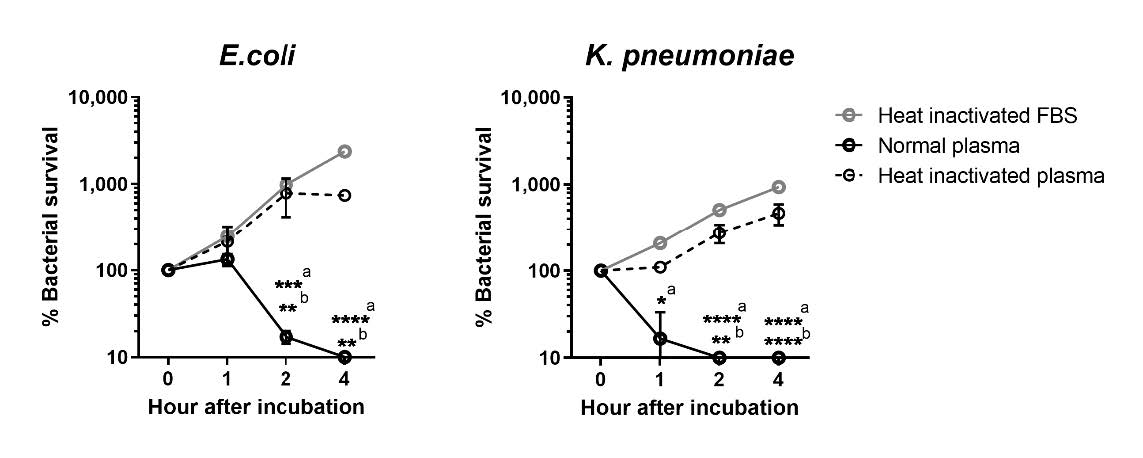
Figure 4. Evaluation of bacteria survival in the presence of normal or heat-inactivated plasma samples. E. coli or K. pneumoniae were cultured in the presence of fetal bovine serum (FBS), normal human plasma or heat-inactivated human plasma for 1, 2 and 4 hours at 37˚C before enumeration of bacterial numbers by standard plating technique on LB agar. Baseline numbers of bacterial before incubation were enumerated as 0 hour after incubation. Data is shown as % bacterial survival calculated from percentage of bacterial number at indicated time point per number of bacteria at baseline. Error bars represent the data from two independent donors. Statistical difference was analyzed by using two-way ANOVA with Tukey's multiple comparisons post-test, *; P < 0.05, **; P < 0.01, ***; P < 0.001, ****; P < 0.0001, a; comparison between normal plasma versus FBS, b; comparison between normal plasma versus heat-inactivated plasma.
DISCUSSION
Bacterial infections are major causes of death, especially in low-income countries (Cars et al. 2005). Thailand provides economic contribution with migrant workers from neighboring countries. The number of Myanmar migrant workers is increasing year by year, since 2012 approximately 1.45 million of Myanmar migrant workers had been working in Thailand (Harkins 2019). Our data showed that whole blood cellular response of IFN- γ response to E. coli, K. pneumoniae, P. aeuroginosa and B. pseudomallei in Myanmar workers were lower than Thai workers. In this study, the lymphocyte counts of Thai people and Myanmar people were not different but Thai people could produce IFN-γ much more than Myanmar people. These data suggest that Myanmar people seem to produce lower cellular effector of IFN-γ response to common bacteria than Thai workers. IFN-γ-mediated immune responses is important in the physiologic response to pathogens by activation of macrophages and stimulation of specific cytotoxic immunity. Cellular immunity mediated by TH1 cells and humoral immunity mediated by TH2 cells are modulated by IFN-γ (Shtrichman et al. 2001). Moreover, the production of IFN-γ may varies with genetic background and ethnic difference can impact cytokine polymorphism (Autenrieth et al. 1994, Govan et al. 2003). Asian people have increased percentage of lower IFN- γ producer genotypes compared to other ethnicities of Whites, Black and Hispanics (Hoffmann et al. 2002). Taken all information along with our data, suggesting that cellular effector of IFN-γ response in Myanmar people seems to be weaker than Thai workers.
Apart from this, antibodies have specific role in defense against infections in participating phagocytosis and complement mediated killing of pathogens. We detected increased antibody level against E. coli, K. pneumoniae, P. aeuroginosa and B. pseudomallei in Myanmar as well as Thai healthy workers suggesting that Myanmar workers might have been already exposed to these common bacteria before and they had developed background IgG levels to these bacteria. In previous experimental infections, host antibody, IgM and IgG can be stimulated by bacteria and these effector antibodies had protected role and clearance of infection (Massironi et al. 2011, Motley et al. 2020, Richard et al. 2020, Chaichana et al. 2021).
Furthermore, B. pseudomallei was observed as the second most common bacteria in hemoculture. This result was expected since the B. pseudomallei infection has been recognized as melioidosis, a lethal infectious disease which is endemic in this area (Limmathurotsakul et al. 2010). Myanmar workers also produced similar level of IgG to B. pseudomallei compared to Thai workers. Seropositivity to B. pseudomallei seemed to be repeated exposure to B. pseudomallei or environment Burkhoderia species (Wuthiekanun et al. 2006, Hantrakun et al. 2018, Rongkard et al. 2020). IgG to low virulent environmental Burkhoderia species of B. thailandensis strains showed cross-protection to B. pseudomallei challenge (Scott et al. 2013). Moreover, antibody to B. pseudomallei did not correlate with duration of staying of Myanmar workers in this endemic area in this study. The presence of background IgG against B. pseudomallei in healthy workers may associate with latent infection or with successful clearance of B. pseudomallei after exposure events or cross-reactive antibody of other Burkhoderia species. Hcp-1 antigen, a diagnosis marker for active melioidosis (Burtnick et al. 2011, Pumpuang et al. 2017) which positivity indicates active melioidosis infection, was applied in this study to exclude the seropositivity against B. pseudomallei from active melioidosis. Our result showed the anti-Hcp-1 antibody was negative, suggesting that Myanmar people may already have been exposed to B. pseudomallei in the past, and they had been developed some type of immunity to this bacterium.
When we investigated the bacteria killing of plasma, by using with low and high level of antibody response to E. coli and K. pneumonia, the plasma could kill the bacteria effectively within hours of infection even having the low level of IgG. This result was consistent with the previous studies of bacterial killing (Strasser et al. 2019). So, humoral response of IgG to common bacteria has a role to kill and clear the bacteria via complement mediated killing.
In conclusion, Thailand now-a-day, is home for Myanmar migrant workers and so healthy social interaction should be promoted to help solve the health risk acquired by migrant workers. Myanmar and Thailand have similar environmental and geographic background and both people are the same ethnic group of Asian. Due to a combination of legal, socio-cultural, behavioral, language and economic barriers migrant workers may have limited awareness or access to health and social services. Providing accessible and quality health services that include prevention, health information. To migrant workers and their families benefits migrant populations.
For safe working of workers, the awareness of health-related risks will be needed to know, and workers should know the information of their health status and preventive measures. We found that Myanmar workers had background immunity against common bacteria in Northeast of Thailand area and Myanmar migrant workers could give similar host response to common risk bacteria in compared with Thai workers even though Myanmar workers shared more or less different cultural adaptation and lifestyle changes. We expect that our study could provide useful health information for Myanmar migrant workers to promote safe working and awareness of health risks to bacterial infections. Social protection in health and increasing social security coverage for Myanmar migrants and their families can enhance access to needed health services and avoid excessive out of pocket payments by migrant workers in need of health services.
ACKNOWLEDGEMENTS
We acknowledge Myanmar and Thai workers for participating in this study and the staff at Department of Cellular and Molecular Immunology KKU for sample collection. M.M.S got scholarship from Graduate School, Khon Kaen University, Thailand.
AUTHOR CONTRIBUTIONS
Myint Myint Sein, Arnone Nithichanon, Chidchamai Kewcharoenwong, and Ganjana Lertmemongkolchai devised and sketched the design of this study. Narisara Chantratita provided and analyzed Hcp-1 protein for this study. Myint Myint Sein analyzed the data and wrote the first draft manuscript. All authors contributed to interpretation of results, reviewed, and approved the submission.
CONFLICT OF INTEREST
The authors declare that they hold no competing interests.
REFERENCES
Abbas, A.K., Lichtman, A.H., Pillai, S., Baker, D.L., and Baker, A. 2018. Cellular and molecular immunology. Philadelphia, PA, Elsevier.
Aung, T.N.N., Shirayama, Y., Moolphate, S., Lorga, T., Yuasa, M., and Nyein Aung, M. 2020. Acculturation and its effects on health risk behaviors among myanmar migrant workers: A cross-sectional survey in Chiang Mai, northern Thailand. International Journal of Environmental Research and Public Health. 17: 5108.
Autenrieth, I.B., Beer, M., Bohn, E., Kaufmann, S.H., and Heesemann, J. 1994. Immune responses to Yersinia enterocolitica in susceptible BALB/c and resistant C57BL/6 mice: An essential role for gamma interferon. Infection and Immunity. 62: 2590-2599.
Brodin, P. and Davis, M.M. 2017. Human immune system variation. Nature Reviews Immunology. 17: 21-29.
Burtnick, M.N., Brett, P.J., Harding, S.V., Ngugi, S.A., Ribot, W.J., Chantratita, N. et al. 2011. The cluster 1 type VI secretion system is a major virulence determinant in Burkholderia pseudomallei. Infection and Immunity. 79: 1512-1525.
Capelli, R., Peri, C., Villa, R., Nithichanon, A., Conchillo-Sole, O., Yero, D. et al. 2018. BPSL1626: Reverse and structural vaccinology reveal a novel candidate for vaccine design against Burkholderia pseudomallei. Antibodies (Basel). 7: 26.
Cars, O. and Nordberg, P. 2005. Antibiotic resistance–The faceless threat. International Journal of Risk & Safety in Medicine. 17: 103-110.
Chaichana, P., Jenjaroen, K., Chumseng, S., Sumonwiriya, M., Rongkard, P., Kronsteiner, B. et al. 2021. Role of Burkholderia pseudomallei-specific IgG2 in adults with acute melioidosis, Thailand. Emerging Infectious Diseases. 27: 463-470.
Chewapreecha, C., Holden, M.T., Vehkala, M., Valimaki, N., Yang, Z., Harris, S.R. et al. 2017. Global and regional dissemination and evolution of Burkholderia pseudomallei. Nature Microbiology. 2: 16263.
Coker, R.J., Hunter, B.M., Rudge, J.W., Liverani, M., and Hanvoravongchai, P. 2011. Emerging infectious diseases in Southeast Asia: Regional challenges to control. Lancet. 377: 599-609.
Cukic, V. and Hadzic, A. 2016. The most common detected bacteria in sputum of patients with community acquired pneumonia (CAP) treated in hospital. Medical Archives. 70: 354-358.
Gourlay, L.J., Peri, C., Ferrer-Navarro, M., Conchillo-Sole, O., Gori, A., Rinchai, D. et al. 2013. Exploiting the Burkholderia pseudomallei acute phase antigen BPSL2765 for structure-based epitope discovery/design in structural vaccinology. Chemical Biology. 20: 1147-1156.
Govan, V.A., Carrara, H.R., Sachs, J.A., Hoffman, M., Stanczuk, G.A., and Williamson, A.L. 2003. Ethnic differences in allelic distribution of IFN-g in South African women but no link with cervical cancer. Journal of Carcinogenesis. 2: 3.
Hantrakun, V., Thaipadungpanit, J., Rongkard, P., Srilohasin, P., Amornchai, P., Langla, S. et al. 2018. Presence of B. thailandensis and B. thailandensis expressing B. pseudomallei-like capsular polysaccharide in Thailand, and their associations with serological response to B. pseudomallei. PLoS Neglected Tropical Diseases. 12: e0006193.
Harkins, B. 2019. Thailand Migration Report 2019. United Nations Thematic Working Group on Migration in Thailand: Bangkok, Thailand.
Hoffmann, S.C., Stanley, E.M., Cox, E.D., DiMercurio, B.S., Koziol, D.E., Harlan, D.M. et al. 2002. Ethnicity greatly influences cytokine gene polymorphism distribution. American Journal of Transplantation. 2: 560-567.
Kak, G., Raza, M., and Tiwari, B.K. 2018. Interferon-gamma (IFN-gamma): Exploring its implications in infectious diseases. Biomolecular Concepts. 9: 64-79.
Kern, W.V. and Rieg, S. 2020. Burden of bacterial bloodstream infection-a brief update on epidemiology and significance of multidrug-resistant pathogens. Clinical Microbiology and Infection. 26: 151-157.
Laupland, K.B., Pasquill, K., Parfitt, E.C., Naidu, P., and Steele, L. 2016. Burden of community-onset bloodstream infections, Western Interior, British Columbia, Canada. Epidemiology and Infection. 144: 2440-2446.
Limmathurotsakul, D., Golding, N., Dance, D.A., Messina, J.P., Pigott, D.M., Moyes, C.L. et al. 2016. Predicted global distribution of Burkholderia pseudomallei and burden of melioidosis. Nature Microbiology. 1: 15008.
Limmathurotsakul, D., Wongratanacheewin, S., Teerawattanasook, N., Wongsuvan, G., Chaisuksant, S., Chetchotisakd, P. et al. 2010. Increasing incidence of human melioidosis in Northeast Thailand. American Journal of Tropical Medicine and Hygiene. 82: 1113-1117.
Liston, A., Carr, E.J., and Linterman, M.A. 2016. Shaping variation in the human immune system. Trends in Immunology. 37: 637-646.
Massironi, S.M., Arslanian, C., Carneiro-Sampaio, M.M., and Pontes, G.N. 2011. Minimal concentration of human IgM and IgG antibodies necessary to protect mice from challenges with live O6 Escherichia coli. FEMS Immunology and Medical Microbiology. 63: 193-201.
Motley, M.P., Diago-Navarro, E., Banerjee, K., Inzerillo, S. and Fries, B.C. 2020. The role of IgG subclass in antibody-mediated protection against carbapenem-resistant Klebsiella pneumoniae. mBio. 11:e02059-20.
Myat, T.O., Oo, K.M., Mone, H.K., Htike, W.W., Biswas, A., Hannaway, R.F. et al. 2020. A prospective study of bloodstream infections among febrile adolescents and adults attending Yangon General Hospital, Yangon, Myanmar. PLOS Neglected Tropical Diseases. 14: e0008268.
Myat, T.O., Prasad, N., Thinn, K.K., Win, K.K., Htike, W.W., Zin, K.N. et al. 2014. Bloodstream infections at a tertiary referral hospital in Yangon, Myanmar. Transactions of the Royal Society of Tropical Medicine and Hygiene. 108: 692-698.
Nahid, P., Jarlsberg, L.G., Kato-Maeda, M., Segal, M.R., Osmond, D.H., Gagneux, S. et al. 2018. Interplay of strain and race/ethnicity in the innate immune response to M. tuberculosis. PLoS One. 13: e0195392.
Nithichanon, A., Rinchai, D., Buddhisa, S., Saenmuang, P., Kewcharoenwong, C., Kessler, B. et al. 2018. Immune control of Burkholderia pseudomallei--common, high-frequency T-cell responses to a broad repertoire of immunoprevalent epitopes. Frontiers in Immunology. 9: 484.
Nithichanon, A., Rinchai, D., Gori, A., Lassaux, P., Peri, C., Conchillio-Sole, O. et al. 2015. Sequence- and structure-based immunoreactive epitope discovery for Burkholderia pseudomallei flagellin. PLoS Neglected Tropical Diseases. 9: e0003917.
Phodha, T., Riewpaiboon, A., Malathum, K., and Coyte, P.C. 2019. Excess annual economic burdens from nosocomial infections caused by multi-drug resistant bacteria in Thailand. Expert Review of Pharmacoeconomics & Outcomes Research. 19: 305-312.
Piasecka, B., Duffy, D., Urrutia, A., Quach, H., Patin, E., Posseme, C. et al. 2018. Distinctive roles of age, sex, and genetics in shaping transcriptional variation of human immune responses to microbial challenges. Proceedings of the National Academy of Sciences of the United States of America. 115: e488-497.
Piruzian, L.A. and Mikhailovskii, E.M. 2009. Metabolic, ethnic, and constitutional specificity of antibacterial immunity. Fiziol Cheloveka. 35: 108-118.
Pumpuang, A., Dunachie, S.J., Phokrai, P., Jenjaroen, K., Sintiprungrat, K., Boonsilp, S. et al. 2017. Comparison of O-polysaccharide and hemolysin co-regulated protein as target antigens for serodiagnosis of melioidosis. PLOS Neglected Tropical Diseases. 11: e0005499.
Richard, G., MacKenzie, C.R., Henry, K.A., Vinogradov, E., Hall, J.C. and Hussack, G. 2020. Antibody binding to the O-specific antigen of Pseudomonas aeruginosa O6 inhibits cell growth. Antimicrobial Agents and Chemotherapy. 64.
Rongkard, P., Kronsteiner, B., Hantrakun, V., Jenjaroen, K., Sumonwiriya, M., Chaichana, P. et al. 2020. Human immune responses to melioidosis and cross-reactivity to low-virulence Burkholderia species, Thailand. Emerging Infectious Diseases. 26: 463-471.
Scott, A.E., Laws, T.R., D'Elia, R.V., Stokes, M.G., Nandi, T., Williamson, E.D. et al. 2013. Protection against experimental melioidosis following immunization with live Burkholderia thailandensis expressing a manno-heptose capsule. Clinical and Vaccine Immunology. 20: 1041-1047.
Shtrichman, R. and Samuel, C.E. 2001. The role of gamma interferon in antimicrobial immunity. Current Opinion in Microbiology. 4: 251-259.
Strasser, J., de Jong, R.N., Beurskens, F.J., Wang, G., Heck, A.J.R., Schuurman, J. et al. 2019. Unraveling the macromolecular pathways of IgG oligomerization and complement activation on antigenic surfaces. Nano Letter. 19: 4787-4796.
van der Maten, E., de Jonge, M.I., de Groot, R., van der Flier, M., and Langereis, J.D. 2017. A versatile assay to determine bacterial and host factors contributing to opsonophagocytotic killing in hirudin-anticoagulated whole blood. Scientific Reports. 7: 42137.
WHO. 2008. Global burden of disease: 2004 update. Geneva, Switzerland. World Health Organization.
Wuthiekanun, V., Chierakul, W., Langa, S., Chaowagul, W., Panpitpat, C., Saipan, P. et al. 2006. Development of antibodies to Burkholderia pseudomallei during childhood in melioidosis-endemic northeast Thailand. American Journal of Tropical Medicine and Hygiene. 74: 1074-1075.
OPEN access freely available online
Natural and Life Sciences Communications
Chiang Mai University, Thailand.
Myint Myint Sein1, 2, Arnone Nithichanon1,3, Narisara Chantratita5, Chidchamai Kewcharoenwong1, 4,* and Ganjana Lertmemongkolchai1,4,*
1 Centre for Research and Development of Medical Diagnostic Laboratories, Faculty of Associated Medical Sciences, Khon Kaen University, Khon Kaen 40000, Thailand.
2 Department of Medical Laboratory Technology, University of Medical Technology, Yangon 11011, Myanmar.
3 Department of Microbiology, Faculty of Medicine, Khon Kaen University, Khon Kaen, 40000, Thailand.
4 Faculty of Associated Medical Sciences, Chiang Mai University, Chiang Mai 50200, Thailand.
5 Department of Microbiology and Immunology, Faculty of Tropical Medicine, Mahidol University, Bangkok 10400, Thailand.
Corresponding author: Ganjana Lertmemongkolchai,
E-mail: ganjana.l@cmu.ac.th; ganja_le@kku.ac.th,
Chidchamai Kewcharoenwong, E-mail: chidchamai.k@cmu.ac.th
Total Article Views
Editor: Veerasak Punyapornwithaya,
Chiang Mai University, Thailand
Article history:
Received: July 5, 2022;
Revised: January 23, 2023;
Accepted: February 2, 2023;
Published online: February 8, 2023

