
Optimization of glucoamylase production by Humicola grisea MTCC 352 in solid state fermentation. Vinayagam Ramesh,* and Vytla Ramachandra Murty
Published Date : 2019-08-23
DOI : https://doi.org/10.12982/CMUJNS.2019.0018
Journal Issues :
Number 3 , July-September 2019
ABSTRACT
In the industrial processing of starch, the thermostable glucoamy- lase is employed in saccharification step. The thermophilic fungi Humicola grisea has been used for the glucoamylase production in solid state fermen- tation. The extracellular glucoamylase is estimated using glucose oxidase – peroxidase assay method. The initial screening studies revealed that wheat bran is the best substrate among the studied agricultural residues. The fer- mentation parameters were optimized through the response surface ap- proach. By using central composite design, the optimal values of four im- portant parameters viz., mineral salt solution concentration, incubation period, initial moisture content and inoculum size for glucoamylase produc- tion were found to be 65 % (v/w), 80 h, 240 % (v/w) and 13 % (v/w) respec- tively. The experimental activity of 282 U/gds obtained was close to the pre- dicted activity of 288 U/gds. A high R2 value (0.9741), P values lesser than 0.05 and AARD values (1.98 %) indicate the accuracy of the proposed model.
Keywords: Glucoamylase, Humicola grisea, Response surface methodology, Solid state fermentation
INTRODUCTION
Glucoamylase (EC 3.2.1.3) is an endo-acting enzyme which cleaves both α-1,4 and α-1,6 linkages of starch and related polymers, capable of converting it completelyintoglucose, when incubated for longerperiods (James and Lee, 1996). One of the vital process requirements for theindustrialapplication of glucoamylase for starch processing is its use at elevated temperatures. Hence, the thermostable glucoamylase is the tool of choice in an industrial setting (Ramesh and Murty, 2014). Of the various reported glucoamylase producers, the thermophilic fun- gus, Humicola grisea, has been proven to be a steady source of thermostable glu- coamylase (Tosi et al., 1993; Campos and Felix, 1995; Ramesh and Murty, 2015).
The conventionalmethod of glucoamylase production employs submerged fermentation (SmF). In the latest industrial practice, glucoamylase production is mainly carried out using solid-state fermentation (SSF) process (Anto et al., 2006). SmF has several disadvantages such as high capital investment, more complex process, less productivity, high water requirement and higher wastewater produc- tion, high energy requirement and high cost for downstream processing (Babu and Satyanarayana, 1995; Pandey, 2003). These issues shift the focus towards SSF process for glucoamylase production. There are dual roles played by the solid substrate used in SSF: nutrient supplement and anchorage for the fungal mycelia.
The best suited substrates for enzyme production in SSF are agricultural waste residues (Ellaiah et al., 2002). There are many agricultural waste residues such as sugarcane bagasse, wheat bran, wheat straw, rice straw, rice bran, maize bran, gram bran and oil cakes, that were successfully utilized for glucoamylase production (Ellaiah et al., 2002; Ramachandran et al., 2004; Bal- kan and Ertan 2007; Bhargav et al., 2008). Apart from agricultural waste resi- dues, cereal flours, waste bread, food wastes, potato residue and tea waste have been effectively used for the production of glucoamylase (Selvakumar et al., 1998; Te Biesebeke et al., 2005; Wang et al., 2009; Lam et al., 2013; Melikoglu et al., 2013). The use of external carbon and nitrogen components along with the solid substrate enhances glucoamylase production (Kunamneni et al, 2005; Prajapati et al, 2013). Mineral salts supplementation is also necessary for glu- coamylase production (Bertolin et al., 2003, Bhatti et al., 2007; Negi et al., 2011).
There are many physico-chemical factors influence glucoamylase pro- duction in SSF such as the type and concentration of solid substrate used, pres- ence of external carbon and nitrogen source, initial moisture content pH of the medium, the age and size of the inoculum, fermentation temperature and dura- tion of fermentation (Baysal et al, 2003; Ramachandran et al., 2004; Couto and Sanromán, 2006; Balkan and Ertan, 2007). Hence, it is necessary to optimize fermentation parameters after identifying the most suitable substrate to enhance glucoamylase production. The influence of the fermentation conditions on glu- coamylase production has been evaluated and reported in several works. Singh and Soni (2001) optimized glucoamylase production by studying different sub- strates, the level and nature of moistening agent, the temperature, the presence or absence of carbon, and nitrogen and mineral supplements. Ellaiah et al., (2002) investigated some factors that influence glucoamylase production in solid state fermentation, including the initial pH and moisture content, the incubation time, the level of salt solution, and the effect of various substrates. Bertolin et al., (2003) investigated the effect of maltose and soluble starch on batch and fed-batch sol- id-state fermentation for glucoamylase production from Aspergillus awamori.
Optimization by the conventional one-variable-at-a-time approach (OVAT) is practiced by keeping all the parameters at a value, while varying a single parameter, at a time. The major disadvantage of OVAT is that it does not include the interaction effects between the variables studied. Also, the net effect of the individual medium constituents on the overall yield is not por- trayed. To overcome these disadvantages, the optimization studies can be performed using statistical techniques such as response surface methodology (RSM). Kumar and Satyanarayana, (2004) and Prajapati et al., (2013) suc- cessfully applied RSM for the production of glucoamylase. In the current in- vestigation, a response surface approach was used for the optimization of enzyme production by SSF. The process variables optimized were incuba- tion time, moisture level, inoculum size and total mineral salt concentration.
MATERIALS AND METHODS
Materials
Commercial quality rice husk and brans of rice, wheat, black gram and maize were obtained from a rural market in Vellore, India. These were used as solid substrate in SSF. Until a consistent weight was achieved, the substrates were oven-dried at 70°C. Glucose oxidase/peroxidase (GOD- POD) assay kit used was obtained from Agappe Diagnostics Ltd (India).
Microorganism and maintenance
The thermophilic fungus, Humicola grisea MTCC 352, was ob- tained from Microbial Type Culture Collection, Chandigarh, India. The strain was grown on Potato Dextrose Agar (PDA) tubes. The slants were kept at 45°C for a 10-day period. The slants were then stored at 4°C, before use.
Inoculum preparation

Enzyme production
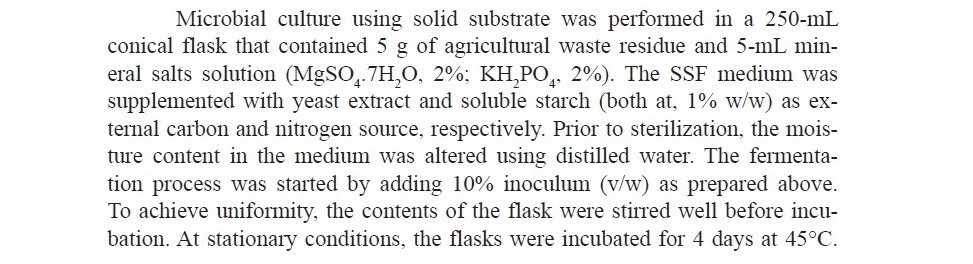
Preparation of crude enzyme
When the fermentation period ended, the entire solid medium was subjected to treat with 50 mL distilled water. This was placed in a shaker that was thoroughly agitated at 100 rpm for a 30-minute period. The suspen- sion was then subjected to filtration using filter paper (Whatman grade 1). The permeate was centrifuged for 10 minutes at a speed of 10,000 rpm, leading to the removal of fungal mycelia. The cell-free supernatant was referred to as crude enzyme and was used throughout the experiments.
Glucoamylase assay
A suitable quantity of crude enzyme was allowed to react with 1% (w/v) soluble starch solution in 50 mM citrate buffer (pH 5.5), at 60°C for the duration of 10 min. The total amount of glucose formed was quantified using Glucose oxidase – peroxidase (GOD-POD) assay kit. A unit of glucoamylase activity is expressed as that quantity of glucoamylase which produces 1 µmole of glucose from starch (soluble) per minute under assay settings.
Central composite design (CCD) and Response surface methodology
![]()
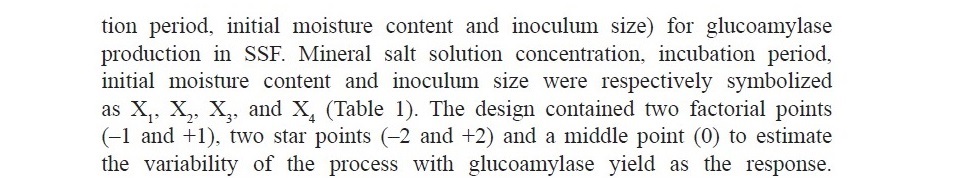
Table 1. Ranges of the independent variables used in central composite design.
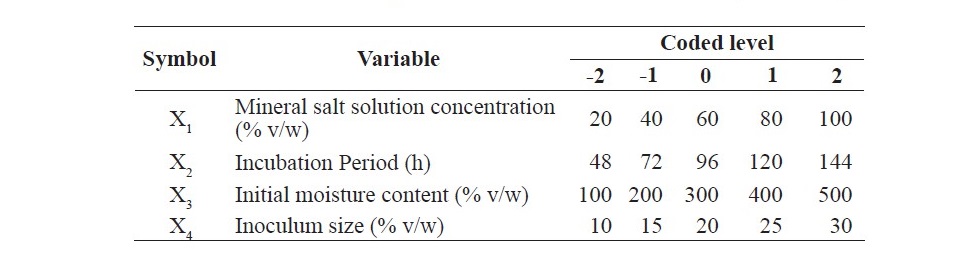
A total of 31 experiments were performed according to the matrix, based on a 4-factor, 5-level CCD (Table 2). The results obtained from experiments were built into a quadratic expression, as a function of the four factors with coded values and is given in equation 1.
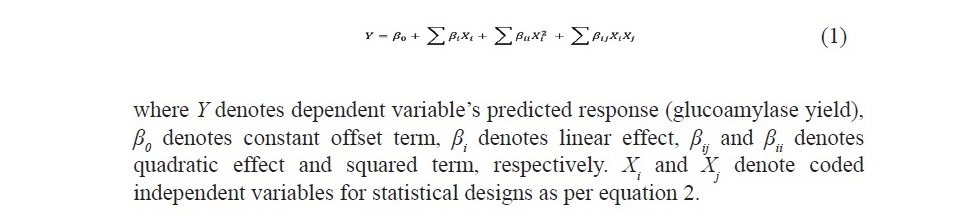
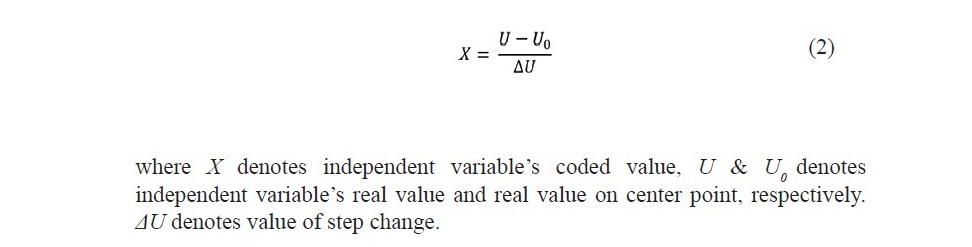
Statistical analysis
Statistical analysis of the model developed by CCD was analyzed by Analysis of variance (ANOVA) concept, by making use of the statistical software package MINITAB-17.1.0 (MINITAB Inc., PA, USA). The polynomial model was statistically verified by using various parameters like linear regres- sion coefficient R2, F- value and absolute average relative deviation (AARD).
RESULTS
Screening of agricultural waste residue for glucoamylase production
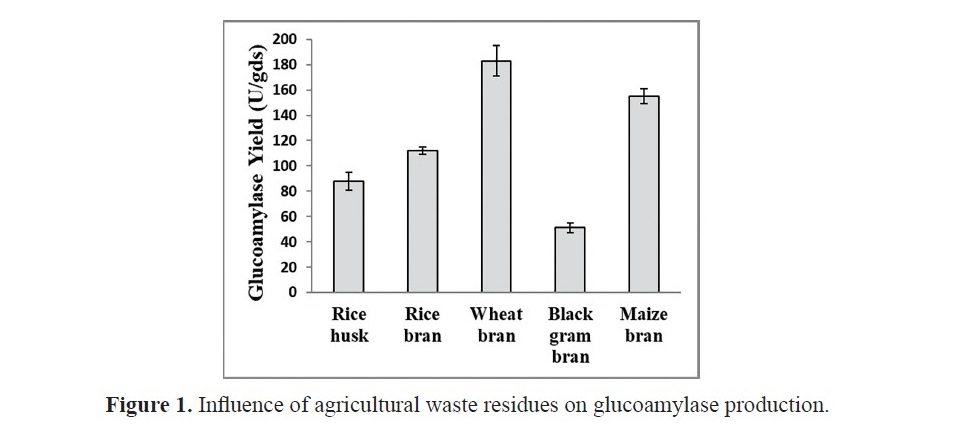
In the present study, five different substrates, viz., rice bran, wheat bran, rice husk, black gram bran & maize bran were tried for extracellular glucoamylase production. Glucoamylase yield of solid state fermentation on various agricultural wastes with 50% of initial moisture is displayed in Figure 1. It was observed that the type of substrates for culturing Humicola grisea played a significant role in the production of glucoamylase. Among the five agricul- tural substrates studied, the maximum glucoamylase yield was obtained with the medium containing wheat bran. On the other hand, lowest enzyme yield was observed with black gram bran. The substrate suitability for glucoamylase production is as per the following order: wheat bran > maize bran > rice bran > rice husk > black gram bran. Thus, wheat bran was selected as the best source for the production of glucoamylase production in the subsequent experiments.
Optimization of fermentation parameters by response surface approach
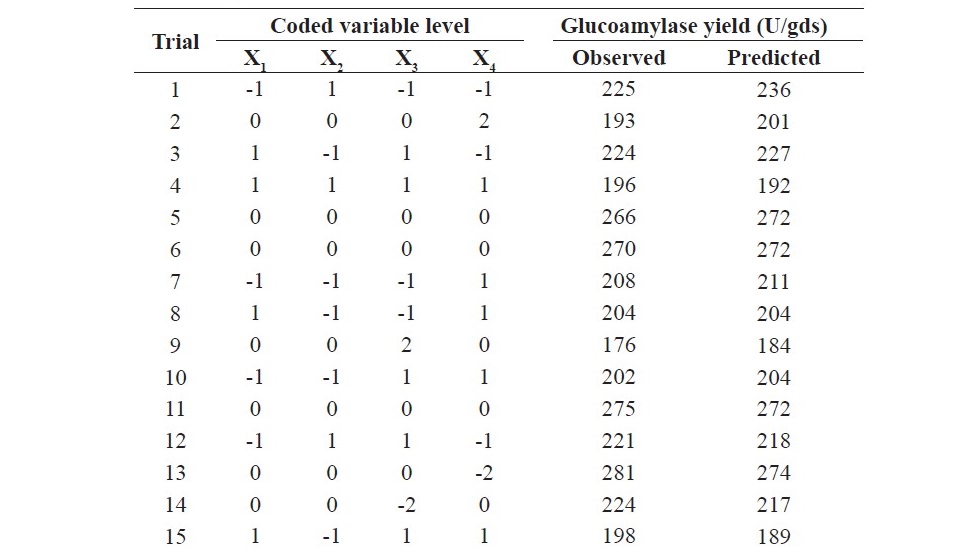
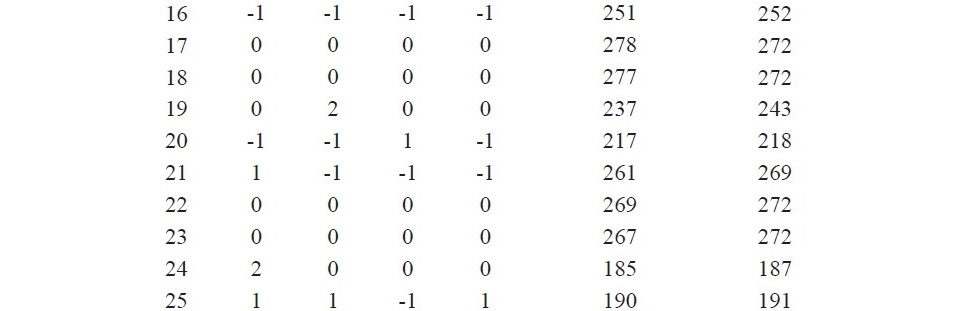
The response surface methodology using CCD was employed for the determination of the optimum value of the four important parameters (min- eral salt solution concentration, incubation period, initial moisture content and inoculum size) for glucoamylase production. A total of 31 experiments were conducted as per the design matrix and the resulting glucoamylase yield is displayed in Table 2 along with the predicted glucoamylase yield.
Table 2. Experimental and predicted responses of the CCD.
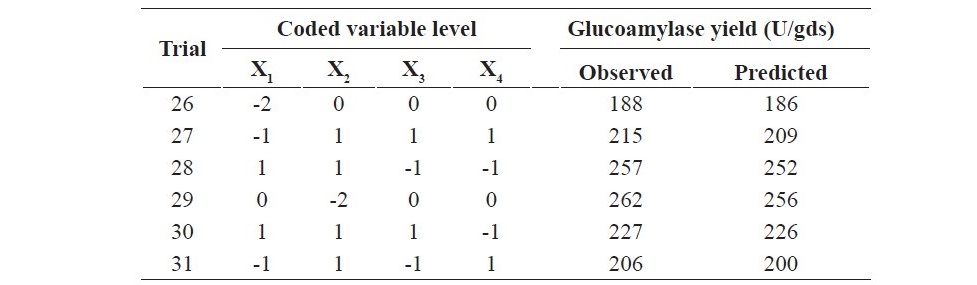
To explain the production of glucoamylase, the second-order regres- sion model equation in terms of coded values was established and expressed in equation 3.
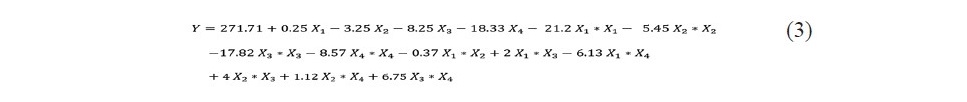
The values of the analysis of variance (ANOVA) for the model are presented in Table 3.
Table 3. ANOVA for the quadratic model.
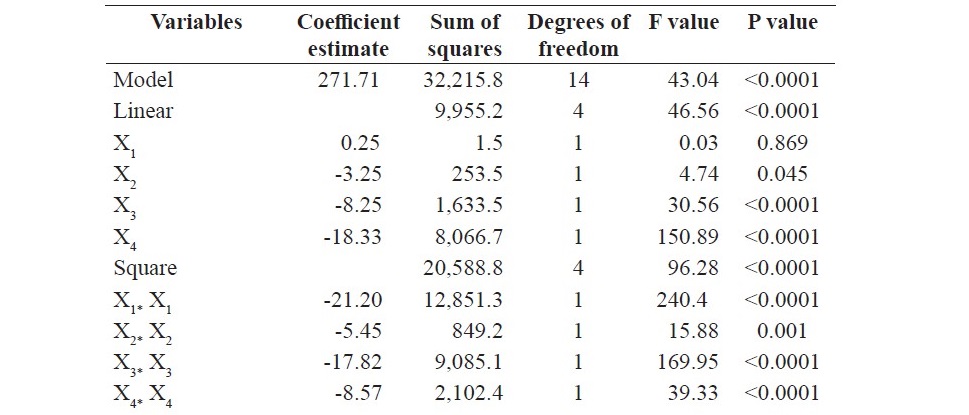
Table 3. Continued
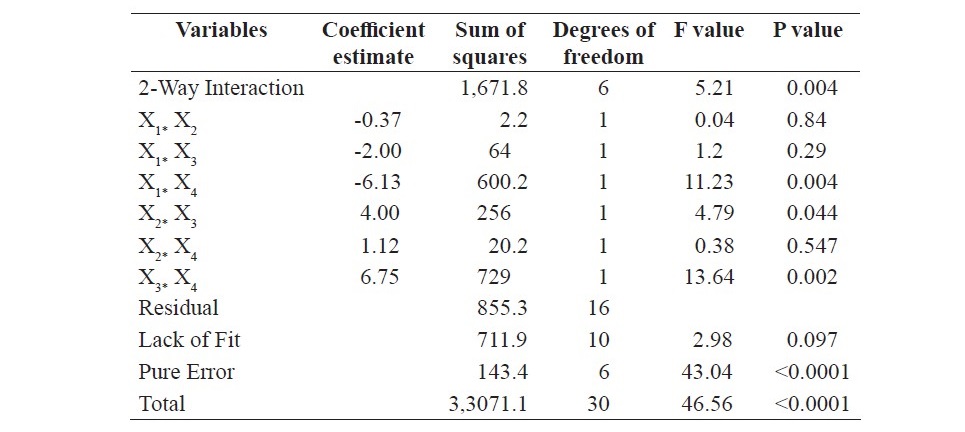
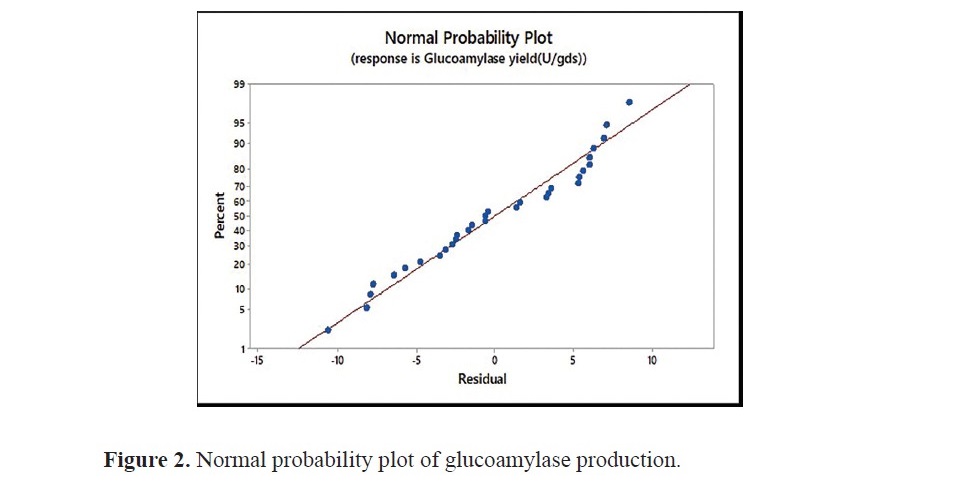
The model resulted in F-value of 43.04 and coefficient of determina- tion (R2): 0. 9741. The lack of fit F value 2.98. The P values were lesser than 0.05 for the linear terms, square effects and interactive effects of mineral salt solution concentration and inoculum size, incubation period and inoculum size as well as initial moisture content and inoculum size. The interaction between the inoculum size and other factors is insignificant for the model obtained. It is clear from the results that the size of inoculum has an independent influence, without interacting with other factors. The P value for the lack of fit was found to be 0.097. For the current system, an AARD of 1.98 % was obtained. Figure 2 displays normal distribution of data as a linear trend, which is the indicator that glucoamylase yield obtained from experiments fits the model equation.
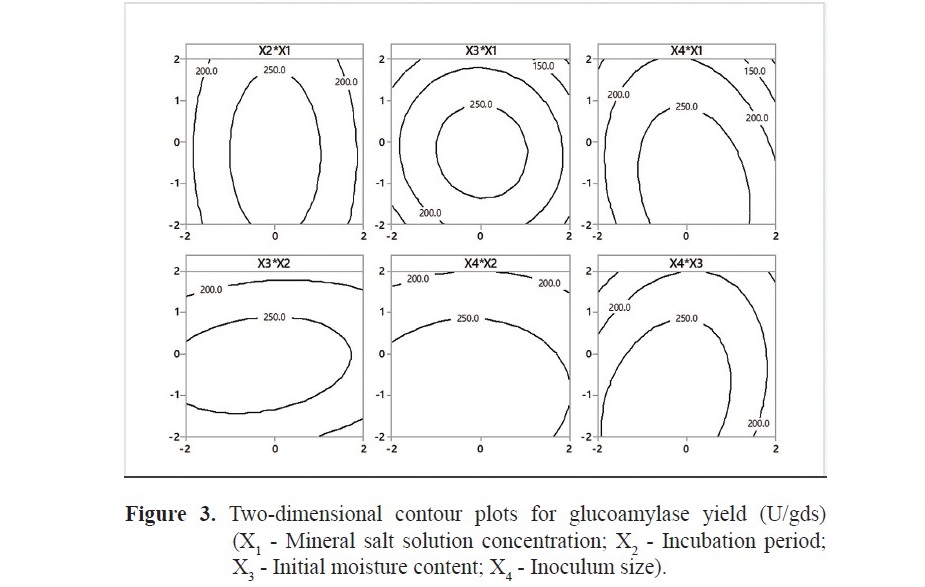
In order to visualize the interaction effects between each variable on glu- coamylase production, two-dimensional contour plots are shown graphically in Figure 3. The interaction effects between two factors are shown with the other two variables kept constant at their center value. It is clear from the plots that there is a change in glucoamylase production with respect to the low or high levels of the factors. The contour plot between the factors, incubation period & initial moisture content indicates the significant interaction effect and an increase in glucoamy- lase production at their higher values. The interaction between mineral salt solu- tion concentration & incubation period and mineral salt solution concentration & initial moisture content shows a negative effect (decrease in glucoamylase pro- duction at higher values). The same phenomena are numerically shown in Table 3.
The response optimizer tool in MINITAB was used to get a solution for the obtained second-order model equation. The optimum levels of each vari- able in uncoded units were as follows: mineral salt solution concentration = 65 % (v/w), incubation period = 80 h, initial moisture content = 240 % (v/w) and inoculum size = 13 % (v/w), all of which were located within the experi- mental range. The predicted glucoamylase yield on wheat bran at the optimum levels of the factors was 288 U/gds. Experiments were performed in triplicates at the optimized values to validate the regression model. Under the optimized conditions, the average of observed experimental values was 282 ± 11 U/gds.
DISCUSSION
The production of extracellular glucoamylase by Humicola grisea MTCC 352 was investigated with numerous readily available agricultural waste residues. The screening is aimed towards the selection of better solid substrate, which is a crucial step in the solid state cultivation for the pro- duction of desired product. Wheat bran, as the most promising solid substrate for glucoamylase production, has been reported by several researchers (Ellaiah et al., 2002; Anto et al., 2006; Bhatti et al., 2007).
The second-order model obtained from the response surface analysis can be sensibly used, as the difference between the obtained and theoretical yield is meagre. The ANOVA results showed higher model F value (43.04) suggests that the second-order model equation obtained was significant. The significance of the second-order model can also be confirmed as results of lack of fit F value. The lower F value of lack of fit (2.98) compared with higher F values of the model suggest the model is significant by means of non-significance lack of fit (Montgomery, 2005). The significance of the second-order regression model can be similarly established with higher coefficient of determination (R2): 0. 9741 in- dicates the extent of correlation between measured glucoamylase yield and model equation. Similarly, the significance of model terms can be established for P val- ues lesser than 0.05 (Montgomery, 2005). The linearity of the normal probability plot confirms all major assumptions of the model viz., distribution of errors, same errors of variance, randomization and mean error stand validated. The AARD explains the extent to which the predicted values differ from the experimental values and a lesser value (<5%) is preferred for a good model (Raja and Murty, 2012). For the current model the AARD value of 1.98 % confirms its adequacy.
Contour plots illustrate the substrate to mineral salt solution concentration and initial moisture content have not supported a higher glucoamylase production at their maximum and minimum levels. Lower mineral salt solution concentration causes insufficient nutrient availability, whereas increase in salt concentration was found to inhibit glucoamylase activity (Kunamneni et al., 2005). The resistance for oxygen transport continuously increases with the decrease in porosity of the agricultural residue resulting from an increase in the moisture level of the solid bed. On the other hand, a decrease in moisture content results in lower solubility of nutrients and reduced availability at microbial surface as well as less swelling of the bed (Ellaiah et al., 2002). A similar effect was observed with incubation period. The incubation period for obtaining the maximum glucoamylase yield is decided based on characteristics of the microorganism and is dependent on the product formation rate. The high inoculum size resulted in the high glucose supplementation to the fungus leading to the decrease in glucoamylase yield.
The good correlation between the observed and predicted glucoamylase yield further confirms the adequacy of the model. In addition to this, the optimized glucoamylase yield was found to be higher than the available literature value for the various microorganisms grown on wheat bran medium such as Aspergillus awamori [13.7 U/gds] (Bertolin et al., 2003), Colletotrichum sp. [61 U/gds] (Prajapati, et al., 2013), Fusarium solani [61.35 U/gds] (Bhatti et al., 2007), Aspergillus awamori [48 U/gds] (Du et al., 2008) and Aspergillus sp [247 U/gds] (Ellaiah et al., 2002).
CONCLUSION
Initial screening study revealed that the type of agricultural waste residue used significantly influences glucoamylase production. Among the tested sources, wheat bran was the best agricultural residue for the glucoamylase production in solid state fermentation. The current study demonstrates the use of response surface approach for optimization of significant fermenta- tion factors which resulted in enhanced glucoamylase yield. The values of the four parameters were optimized by employing CCD (mineral salt solution concentration: 65 % (v/w), incubation period: 80 h, initial moisture content: 240 % (v/w) and inoculum size: 13 % (v/w)). The proposed second-or- der model was validated as the difference between the obtained experimental glucoamylase yield of 282 ± 11 U/gds and the predicted glucoamylase yield of 288 U/gds, which was meagre. Thus, the optimized conditions for the solid state fermentation found out in the current study might reduce the overall cost of the production and provides a basis for further studies on a large scale.
ACKNOWLEDGEMENTS
The authors gratefully acknowledge the Department of Biotechnology, MIT, Manipal Academy of Higher Education for providing the facilities to carry out the research work.
REFERENCES
Anto, H., Trivedi, U.B., and Patel, K.C. 2006. Glucoamylase production by solid-state fermentation using rice flake manufacturing waste products as substrate. Bioresource Technology. 97(10): 1161-1166. https://doi. org/10.1016/j.biortech.2005.05.007
Babu, K.R., and Satyanarayana, T. 1995. α-Amylase production by thermophilic Bacillus coagulans in solid state fermentation. Process Biochemistry. 30(4): 305-309. https://doi.org/10.1016/0032-9592(95)87038-5
Balkan, B., and Ertan, F. 2007. Production of α-Amylase from Penicillium chrysogenum under solid-state fermentation by using some agricultural by-products. Food Technology and Biotechnology. 45(4): 439-442.
Baysal, Z., Uyar, F., and Aytekin, Ç. 2003. Solid state fermentation for produc- tion of α-amylase by a thermotolerant Bacillus subtilis from hot-spring water. Process Biochemistry. 38(12): 1665-1668. https://doi.org/10.1016/ S0032-9592(02)00150-4
Bertolin, T.E., Schmidell, W., Maiorano, A.E., Casara, J., and Costa, J.A. 2003. Influence of carbon, nitrogen and phosphorous sources on glucoamylase production by Aspergillus awamori in solid state fermentation. Zeitschrift für Naturforschung C. 58(9-10): 708-712. https://doi.org/10.1515/znc-2003-9-1020
Bhargav, S., Panda, B.P., Ali, M., and Javed, S. 2008. Solid-state fermentation: an overview. Chemical and Biochemical Engineering Quarterly. 22(1): 49-70.
Bhatti, H.N., Rashid, M.H., Nawaz, R., Asgher, M., Perveen, R., and Jabbar, A. 2007. Optimization of media for enhanced glucoamylase production in solid-state fermentation by Fusarium solani. Food Technology and Biotechnology. 45(1): 51-56.
Campos, L., and Felix, C.R. 1995. Purification and characterization of a glucoamylase from Humicola grisea. Applied and Environmental Micro- biology. 61(6): 2436-2438.
Couto, S.R., and Sanromán, M.A. 2006. Application of solid-state fermentation to food industry—a review. Journal of Food Engineering. 76(3): 291- 302. https://doi.org/10.1016/j.jfoodeng.2005.05.022
Du, C., Lin, S.K.C., Koutinas, A., Wang, R., Dorado, P., and Webb, C. 2008. A wheat biorefining strategy based on solid-state fermentation for fer- mentative production of succinic acid. Bioresource Technology. 99(17): 8310-8315. https://doi.org/10.1016/j.biortech.2008.03.019
Ellaiah, P., Adinarayana, K., Bhavani, Y., Padmaja, P., and Srinivasulu, B. 2002. Optimization of process parameters for glucoamylase production under solid state fermentation by a newly isolated Aspergillus species. Process Biochemistry. 38(4): 615-620. https://doi.org/10.1016/S0032-9592(02)00188-7
James, J.A., and Lee, B.H. 1996. Characterization of glucoamylase from Lactobacillus amylovorus ATCC 33621. Biotechnology Letters. 18(12):1401-1406. https://doi.org/10.1007/BF00129343
Kumar, S., and Satyanarayana, T. 2004. Statistical optimization of a thermo- stable and neutral glucoamylase production by a thermophilic mold Thermomucor indicae-seudaticae in solid-state fermentation. World Journal of Microbiology and Biotechnology. 20(9): 895-902. https://doi. org/10.1007/s11274-004-2891-z
Kunamneni, A., Permaul, K., and Singh, S. 2005. Amylase production in solid state fermentation by the thermophilic fungus Thermomyces lanuginsus. Journal of Bioscience and Bioengineering. 100(2): 168-171. https://doi. org/10.1263/jbb.100.168
Lam, W.C., Pleissner, D., and Lin, C.S.K. 2013. Production of fungal glucoam- ylase for glucose production from food waste. Biomolecules. 3(3): 651- 661. https://doi.org/10.3390/biom3030651
Melikoglu, M., Lin, C.S.K., and Webb, C. 2013. Stepwise optimisation of enzyme production in solid state fermentation of waste bread pieces. Food and Bioproducts Processing. 91(4): 638-646. https://doi.org/10.1016/j. fbp.2013.04.008
Montgomery, D.C. 2005. Design and analysis of experiments. John Wiley & Sons.
Negi, S., Gupta, S., and Banerjee, R. 2011. Extraction and purification of glucoamylase and protease produced by Aspergillus awamori in a single-stage fermentation. Food Technology and Biotechnology. 49(3): 310-315.
Pandey, A. 2003 . Solid-state fermentation. Biochemical Engineering Journal.
13(2): 81-84. https://doi.org/10.1016/S1369-703X(02)00121-3
Prajapati, V.S., Trivedi, U.B., and Patel, K.C. 2013. Optimization of glucoam- ylase production by Colletotrichum sp. KCP1 using statistical meth- odology. Food Science and Biotechnology. 22(1): 31-38. https://doi. org/10.1007/s10068-013-0005-0
Raja, S., and Murty, V.R. 2012. Development and evaluation of environmentally benign aqueous two phase systems for the recovery of proteins from tannery waste water. ISRN Chemical Engineering. 2012: 1-9. http://dx. doi.org/10.5402/2012/290471
Ramachandran, S., Patel, A.K., Nampoothiri, K.M., Francis, F., Nagy, V., Szakacs, G., and Pandey, A. 2004. Coconut oil cake––a potential raw material for the production of α-amylase. Bioresource Technology. 93(2): 169-174. https://doi.org/10.1016/j.biortech.2003.10.021
Ramesh, V., and Murty, V.R. 2014. Sequential statistical optimization of media components for the production of glucoamylase by thermophilic fungus Humicola grisea MTCC 352. Enzyme Research. 2014: 1-9. https://doi. org/10.1155/2014/317940
Ramesh, V., and Murty, V.R. 2015. Partitioning of thermostable glucoamylase in polyethyleneglycol/salt aqueous two-phase system. Bioresources and Bioprocessing. 2(1): 1-8. https://doi.org/10.1186/s40643-015-0056-6
Selvakumar, P., Ashakumary, L., and Pandey, A. 1998. Biosynthesis of glucoam- ylase from Aspergillus niger by solid-state fermentation using tea waste as the basis of a solid substrate. Bioresource Technology. 65(1): 83-85. https://doi.org/10.1016/S0960-8524(98)00012-1
Singh, H., and Soni, S.K. 2001. Production of starch-gel digesting amylo- glucosidase by Aspergillus oryzae HS-3 in solid state fermentation. Process Biochemistry. 37(5): 453-459. https://doi.org/10.1016/S0032-9592(01)00238-2
Te Biesebeke, R., Record, E., Van Biezen, N., Heerikhuisen, M., Franken, A., Punt, P.J., and Van Den Hondel, C.A.M.J.J. 2005. Branching mutants of Aspergillus oryzae with improved amylase and protease production on solid substrates. Applied Microbiology and Biotechnology. 69(1): 44-50. https://doi.org/10.1007/s00253-005-1968-4
Tosi, L.R.O., Terenzi, H.F., and Jorge, J.A. 1993. Purification and characteri- zation of an extracellular glucoamylase from the thermophilic fungus Humicola grisea var. thermoidea. Canadian Journal of Microbiology. 39(9): 846-852. https://doi.org/10.1139/m93-126
Wang, R., Godoy, L.C., Shaarani, S.M., Melikoglu, M., Koutinas, A., and Webb, C. 2009. Improving wheat flour hydrolysis by an enzyme mixture from solid state fungal fermentation. Enzyme and Microbial Technology. 44(4): 223-228. https://doi.org/10.1016/j.enzmictec.2008.10.002
Vinayagam Ramesh1* and Vytla Ramachandra Murty2
1Department of Chemical Engineering, Manipal Institute of Technology, Manipal Academy of Higher Education, Manipal 576104, Karnataka, India
2Department of Biotechnology, Manipal Institute of Technology, Manipal Academy of Higher Education, Manipal 576104, Karnataka, India
*Corresponding author. E-mail: rameshvinayagam@gmail.com
Total Article Views
Article history:
Received: July 16, 2018;
Revised: October 25, 2018;
Accepted: October 29, 2018

