
Detecting Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) and Bacteria on Highly Common Contaminated Surfaces at Urban Hospital and Public Areas
Nattamon Niyomdecha*, Pirom Noisumdaeng, Pakkaporn Archawametheekul, Sunisata Angkham, Benja Norapong, Muttika Fungkrajai, and Pramote SriwanitchrakPublished Date : 2023-01-05
DOI : https://doi.org/10.12982/NLSC.2023.010
Journal Issues : Number 1, January-March 2023
Abstract The persistence of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) on surfaces in public areas increases a risk of Coronavirus disease 2019 (COVID-19) transmission. Alcohol products have been widely used to eliminate virus contaminants. Long-term microbial exposure to alcohol during pandemic may lead to an alcoholic tolerance emerging. We aimed to detect SARS-CoV-2 and bacteria on highly contaminated surfaces at hospital and public areas in Pathum Thani province during the late of 2021, and to evaluate alcohol susceptibility of sampled bacterial isolates. Fifty pooled swab samples were collected from environmental surfaces. Each sample was divided into two parts to test for SARS-CoV-2 and bacteria, respectively. Molecular assay was used to detect SARS-CoV-2, while bacterial detection was done by traditional culture method. SARS-CoV-2 was detected at 8% (4/50) from hospital restroom doorknobs and faucets, as well as on ATM keypads at the center market. Conversely, 96% (48/50) of bacterial contaminants were detected. Of these, only 25 isolates of medically important bacteria were identified and classified into three groups including Staphylococcus, Enterobacteriaceae and non-fermentative bacteria. All isolates were then performed an alcohol susceptibility test with three different alcohol formulas (70% ethanol, 80% ethanol and 75% isopropanol), Results were compared with standard strains (ATCC) and isolated bacteria obtained from before the COVID-19 pandemic. Isolated bacteria from this study were significantly less susceptible to all types of alcohol tested, and Staphylococcus was the most resistant to destruction. Thus, an effective infection control measure in a high-risk area should be considered and regularly practiced.
Keywords: SARS-CoV-2, COVID-19, Bacteria, Highly contaminated surfaces, Alcohol susceptibility test
Citation: Niyomdecha, N., Noisumdaeng, P., Archawametheekul, P., Angkham, S., Norapong, B., Fungkrajai, M., and Sriwanitchrak, P. 2023. Detecting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and bacteria on highly common contaminated surfaces at urban hospital and public areas. Nat. Life Sci. Commun. 22(1): e2023010.
INTRODUCTION
Rapid spreading infection across the world of the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) leads to the Coronavirus disease 2019 (COVID-19) pandemic. Exposures to infectious SARS-CoV-2 respiratory droplets can occur via direct contact from person to person and indirect contact involving touching with contaminated fomites that virus can enter to susceptible mucous membranes (Castano et al., 2021). Available data are still controversial regarding the significance of fomite-mediated transmission (Mohamadi et al., 2021; Pastorino et al., 2020). Although an indirect transmission may have a lower risk to cause disease, prolonged persistence of contaminated surfaces with viable SARS-CoV-2 (Cervino et al., 2020) should be awareness and tracked.
In response to the COVID-19 pandemic, keeping hand hygiene using soap and water or alcohol-based hand sanitizers (ABHSs) plays a crucial role in mitigating SARS-CoV-2 transmission and infection (Assefa and Melaku, 2021). The Food and Drug Administration (FDA) has recommended sanitizing of non-visibly soiled hands with an alcohol-based formulation agent containing 80% v/v ethanol or 75% v/v isopropanol (FDA, 2020). An overwhelming increase in hand sanitizer demand contributed to a substantial shortage of supply resulting in the acceleration of productions. The number of commercially available sanitizers are produced with less rigorous quality control. Frequently exposure to substandard ABHS products and inappropriate use can raise the problems of safety concerns. The great impacts may occur from a progressive stepwise accumulation of natural mutations involving alcohol resistance gene in microorganisms (Assefa and Melaku, 2021; Mahmood et al., 2020; Pidot et al., 2018).
Thus, this study aimed to detect the genetic material RNA of SARS-CoV-2 and bacteria from environmental samples in the public areas of hospital, university, and community sites. Additionally, we determined the alcohol susceptibility of medically important bacterial isolates derived between before and after COVID-19 pandemic. This provides a prevalence of SARS-CoV-2 and bacteria in highly touched environmental surfaces and an insight data regarding the emerging of alcoholic tolerance in bacteria.
MATERIALS AND METHODS
Approval statement and study sites
All procedures in this project were approved by the Thammasat University Institutional Biosafety Committee (101/2564). All handing of suspected SARS-CoV-2 environmental samples was performed under biosafety level 2 plus (BSL-2 plus) in accordance with the biosafety guidelines.
During October to November 2021, swab samples collected from highly touched environmental surfaces was conducted in public areas of Thammasat University, Thammasat University Hospital, and nearby public places at Pathum Thani province, Thailand. Pathum Thani province, located at the central part of Thailand, was the one of 29 provinces that had been designated as the Maximum and Strict Controlled Areas or dark-red zone provinces since the mid-year of 2021.
Evaluation of sample collection process
Since this study used a wooden cotton-tipped swab to collect the samples that might have a negative impact on the PCR reaction due to its structure components or any impurity substances; thus, we initially evaluated the recovery result of SARS-CoV-2 RNA using RT-qPCR detection on artificial swab samples. Forty microliters (µl) of SARS-CoV-2 RNA (50 ng/µl) were randomly spotted onto aluminum foil and sterile petri dish within area of 10 x 10 cm and let it dry. Sterile cotton swab was then used to swab the entire surface, and swab was resuspended in 200 µl of nuclease-free water to make a concentration of 10 ng/µl.
Furthermore, our study designed to collect samples from each indicated site by pooling swab process instead of single swab. Likewise, we also conducted the artificial spiked-SARS-CoV-2 RNA in negative SARS-CoV-2 detected pool swab sample. Briefly, 40 µl of SARS-CoV-2 RNA (50 ng/µl) was spiked in negative pooled swab sample to make a final concentration of 10 ng/µl. These artificial swab samples were analyzed and compared the RT-qPCR result with SARS-CoV-2 RNA template at same concentration.
Environmental sampling
Fifty pooled swab samples collected from high risk-contaminated SARS-CoV-2 surfaces were taken to analyze. Sampling was done in main critical areas which located outside inhabited sites of definite COVID-19 cases, including cafeterias, toilets, public spaces, corridors between buildings, and public shuttle bus. Various sampled objects were selected to swab their surfaces as shown in Table 2.
Sterile cotton-tipped applicator was moistened with nuclease-free water. Surface objects were swabbed in an area of 10 x 10 cm. or entire area for smaller ones. Ten swabs were used to randomly swab in each defined object, and they were resuspended in 700 µl of nuclease-free water. Pooled swab samples were maintained at 4-8°C throughout transportation, and sample processing was performed within 2-3 h after collection. All processes were done under the SARS-CoV-2 biosafety guidelines, and suspected infectious materials were decontaminated and discarded properly.
RNA extraction and RT-qPCR test
Exact volume of samples was aliquoted to extract RNA by using TRIzolTM LS (Invitrogen)-chloroform method or QIAamp Viral RNA Mini Kit (QIAGEN, Cat no. 52904) according to manufacturer’s guidelines. RNA was eluted in 20-50 µl of RNase-free water, and purified RNA was measured a concentration as ng/µl and kept at -80°C.
A Luna® Universal One-Step RT-qPCR Kit (New England Biolabs) was used for RT-qPCR to detect SARS-CoV-2 targeting N gene with specific primers. Primer sequences were previously designed from the Charite/Berlin protocol (Corman et al., 2020) as follows: N_Sarbeco_F: CACATTGGCACCCGCAATC; N_Sarbeco_R: GAGGAACGAGAAGAGGCTTG. Final reaction mixture included 1X one-step reaction mix, 1X warmstart RT enzyme mix, 200 nM of each primer, 50 ng of RNA template, and nuclease-free water used for volume adjustment equal to 20 µl. SARS-CoV-2 RNA (kindly provided by Professor Prasert Auewarakul, Mahidol University) and nuclease-free water were used as positive and negative controls respectively. Thermal cycling was performed by using CFX-96 real-time PCR detection system (Bio-Rad, USA) at 55°C for 10 min for reverse transcription, followed by 95°C for 1 min, and then 40 cycles of 95°C for 10 s, 60°C for 30 s, and melt curve analysis.
Cultivation and identification of bacteria
Three hundred µl of swab samples in nuclease-free water was transferred into 4 ml of nutrient broth, and culture medium were incubated at 37°C for 16-18 h. Turbidity was observed, and a full-loop of turbid medium was inoculated on blood agar and MacConkey agar. Culture plates were incubated at 37°C for 16-18 h. Different colonies in morphology were picked and processed by Gram stain, presumptive test, and biochemical tests. Results were then recorded and interpreted, and only medically important bacteria strains that can be identified by biochemical tests were selected to subculture for subsequent experiment.
Alcohol susceptibility test
Alcohol susceptibility test was conducted with selected bacteria isolate as described above. The results were compared with bacteria derived before COVID-19 pandemic from ATCC (kindly obtained from Department of Medical Technology, Thammasat University) and clinical specimens (kindly provided by Dr. Worada Samosornsuk and Dr. Anek Pootong at Thammasat University).
A standard antimicrobial assay of spot plating was used as described elsewhere (Chojnacki et al., 2021). Four to five of bacterial isolated colonies were randomly taken from pure culture and resuspended in sterile normal saline to adjust the turbidity equal to 0.5 McFarland standard. In duplicate, non-toxic and sterile swab was used to transfer the inoculum onto Mueller-Hinton agar (MHA). Swab was dipped and the excess liquid was removed by turning the swab against the side of the tube. The entire surface of MHA was spread by swabbing in three directions and dried for 5 min before applying alcohol reagent tested. Twenty-five µl of each different alcohol formulation, including 70% ethanol, 80% ethanol, and 75% isopropanol (IPA), was spotted onto the agar surface, and then incubated at 37°C for 16-18 h. Equal volume of sterile nuclease-free water was also used to spot on plate as negative control. The diameter of inhibition zone was measured in millimeters (mm), and average zone of inhibition (ZOI) and standard deviation (SD) were recorded. Colonies growing within the ZOI were counted and recorded to indicate the potential for bacterial tolerance to each alcohol type.
Statistical analysis
Data are described herein as the mean ± SD. Statistical analysis was performed by using unpaired Student's t-tests for comparing the differences between groups. Significance was accepted when P < 0.05.
RESULTS
Evaluation of the recovery result of SARS-CoV-2 detection method from artificial samples
Since a collection of samples with cotton swab and pooled swab assay may have a negative effect on RT-qPCR reaction, the recovery result of SARS-CoV-2 detection from artificially contaminated samples was primarily evaluated. As shown in Table 1, spiked SARS-CoV-2 RNA could be detected from artificial samples, although cycle threshold (Ct) variation was found when compared to unspiked RNA. Averages of Ct variation value from using single cotton swab to collect spiked RNA on artificial material surfaces and from artificially spiked RNA in negative for SARS-CoV-2 pooled swab samples were 3.3 and 7.8, respectively.
Table 1. The recovery of SARS-CoV-2 detection method on artificial samples.
|
Tested samples with |
Cycle threshold (Ct) values |
Average Ct |
|
|
1 |
2 |
|
|
|
Unspiked (PC) |
19.49 |
19.14 |
19.32 |
|
Spiked on aluminum foil |
22.88 |
22.94 |
22.91 |
|
Spiked on plastic dish |
22.10 |
22.56 |
22.33 |
|
Spiked in pooled swab sample |
27.06 |
27.25 |
27.16 |
Note: Abbreviations: PC, Positive control.
SARS-CoV-2 detection from environmental samples
As shown in Table 2, 50 pooled swab samples were collected from various kinds of commonly touched surface objects in public areas of non-inhabited sites of COVID-19 patients in hospital, University and community places. The positive SARS-CoV-2 detection rate was at 8% (4/50). The highest risk area possessing the highest accumulative positive results was at hospital’s toilet (3/15), which SARS-CoV-2 RNA was detected at faucet handles and doorknobs/door latchs. Additionally, SARS-CoV-2 RNA could be found on keypad of Automatic Teller Machine (ATM) at the center market (1/5). Positive Ct values from these surfaces were 34.06, 35.14, 34.96, and 38.24, respectively.
Bacteria detection and identification of medically important bacteria from environmental samples
Positive rate of bacteria detection by culture method from same objected samples was dramatically higher than SARS-CoV-2 detection. In Table 3, contaminated bacteria were found on almost surface objects (48/50, 96%). However, major colony appearance on agar and gram stain were Bacillus spp., which most species are non-pathogenic strains that can be commonly found in environments. Indeed, only 25 medically important bacteria isolates were identified with biochemical tests as shown in Table 4. Of which, 6 isolates were Staphylococcus spp., 10 isolates were bacteria in Enterobacteriaceae, and 9 isolates were non-fermentative bacteria.
Table 2. SARS-CoV-2 detection from environmental samples
|
Places and Areas |
No. of RNA-positive samples/total no. of samples (%) |
Total (%) |
||||||||
|
Doorknob |
Faucet handle |
Toilet seat |
Toilet flush |
Table and chair |
Divider screen |
Handrail |
Keypad of ATM |
Elevator button |
||
|
At hospital: |
|
|
|
|
|
|
|
|
|
|
|
Cafeteria |
- |
- |
- |
- |
0/1 (0%) |
0/1 (0%) |
0/1 (0%) |
- |
- |
0/3 (0%) |
|
Cafeteria’s toilet |
0/2 (0%) |
0/2 (0%) |
- |
- |
- |
- |
- |
- |
- |
0/4 (0%) |
|
Hospital’s toilet |
1/5 (20%) |
2/5 (40%) |
0/3 (0%) |
0/2 (0%) |
- |
- |
- |
- |
- |
3/15 (20%) |
|
Public space |
- |
- |
- |
- |
0/1 (0%) |
- |
0/1 (0%) |
0/2 (0%) |
0/2 (0%) |
0/6 (0%) |
|
Corridor between buildings |
- |
- |
- |
- |
- |
- |
- |
0/1 (0%) |
- |
0/1 (0%) |
|
At University: |
|
|
|
|
|
|
|
|
|
|
|
Public space |
- |
- |
- |
- |
0/1 (0%) |
- |
0/1 (0%) |
- |
0/2 (0%) |
0/4 (0%) |
|
Cafeteria's toilet |
0/1 (0%) |
0/1 (0%) |
- |
- |
- |
- |
- |
- |
- |
0/2 (0%) |
|
Gymnasium |
- |
- |
- |
- |
0/1 (0%) |
- |
- |
- |
- |
0/1 (0%) |
|
Gymnasium's toilet |
0/1 (0%) |
0/1 (0%) |
- |
- |
- |
- |
- |
- |
- |
0/2 (0%) |
|
Shuttle bus |
- |
- |
- |
- |
- |
- |
0/1 (0%) |
- |
- |
0/1 (0%) |
|
At center market: |
|
|
|
|
|
|
|
|
|
|
|
Public space |
- |
- |
- |
- |
- |
- |
- |
1/5 (20%)
|
- |
1/5 (20%) |
|
At petrol stations: |
|
|
|
|
|
|
|
|
|
|
|
Toilet |
0/2 (0%) |
0/2 (0%) |
- |
- |
- |
- |
- |
- |
- |
0/4 (0%) |
|
Public space |
- |
- |
- |
- |
- |
- |
- |
0/2 (0%) |
- |
0/2 (0%) |
|
Total (%) |
1/11 (9.0%) |
2/11 (18.2%) |
0/3 (0%) |
0/2 (0%) |
0/4 (0%) |
0/1 (0%) |
0/4 (0%) |
1/10 (10%) |
0/4 (0%) |
4/50 (8%) |
Table 3. Bacteria detection from environmental samples.
|
Places and Areas |
No. of RNA-positive samples/total No. of samples (%) |
Total (%) |
|
|||||||||
|
Doorknob |
Faucet handle |
Toilet seat |
Toilet flush |
Table and chair |
Divider screen |
Handrail |
Keypad of ATM |
Elevator button |
|
|||
|
At hospital:: |
|
|
|
|
|
|
|
|
|
|
||
|
Cafeteria |
- |
- |
- |
- |
1/1 (100%) |
1/1 (100%) |
1/1 (100%) |
- |
- |
3/3 (100%) |
||
|
Cafeteria’s toilet |
2/2 (100%) |
2/2 (100%) |
- |
- |
- |
- |
- |
- |
- |
4/4 (100%) |
||
|
Hospital’s toilet |
5/5 (100%) |
5/5 (100%) |
3/3 (100%) |
2/2 (100%) |
- |
- |
- |
- |
- |
15/15 (100%) |
||
|
Public space |
- |
- |
- |
- |
1/1 (100%) |
|
1/1 (100%) |
2/2 (100%) |
2/2 (100%) |
6/6 (100%) |
||
|
Corridor between buildings |
- |
- |
- |
- |
- |
- |
- |
0/1 (0%) |
- |
0/1 (0%) |
||
|
At Universit: |
|
|
|
|
|
|
|
|
|
|
||
|
Public space |
- |
- |
- |
- |
1/1 (100%) |
- |
1/1 (100%) |
- |
2/2 (100%) |
4/4 (100%) |
||
|
Cafeteria's toilet |
1/1 (100%) |
1/1 (100%) |
- |
- |
- |
- |
- |
- |
- |
2/2 (100%) |
||
|
Gymnasium |
- |
- |
- |
- |
1/1 (100%) |
- |
- |
- |
- |
1/1 (100%) |
||
|
Gymnasiu’s toilet |
1/1 (100%) |
1/1 (100%) |
- |
- |
- |
- |
- |
- |
- |
2/2 (100%) |
||
|
Shuttle bus |
- |
- |
- |
- |
- |
- |
1/1 (100%) |
- |
- |
1/1 (100%) |
||
|
At center market: |
|
|
|
|
|
|
|
|
|
|
||
|
Public space |
- |
- |
- |
- |
- |
- |
- |
5/5 (100%) |
- |
5/5 (100%) |
||
|
At petrol stations: |
|
|
|
|
|
|
|
|
|
|
||
|
Toilet |
2/2 (100%) |
2/2 (100%) |
- |
- |
- |
- |
- |
- |
- |
4/4 (100%) |
||
|
Public space |
- |
- |
- |
- |
- |
- |
- |
1/2 (50%) |
- |
1/2 (50%) |
||
|
Total (%) |
11/11 (100%) |
11/11 (100%) |
3/3 (100%) |
2/2 (100%) |
4/4 (100%) |
1/1 (100%) |
4/4 (100%) |
8/10 (100%) |
4/4 (100%) |
48/50 (96%) |
||
Table 4. Identification of medically important bacteria from environmental samples.
|
Places and Areas |
Objects |
No. of isolates |
||||||
|
S. aureus |
CoNS |
E. coli |
K. pneumoniae |
P. aeruginosa |
A. baumannii |
Total |
||
|
At hospital: |
|
|
|
|
|
|
|
|
|
Cafeteria |
Handrail |
1 |
|
|
|
|
|
1 |
|
Cafeteria's toilet |
Faucet handle |
|
|
2 |
1 |
1 |
1 |
5 |
|
|
Doorknob |
1 |
|
1 |
1 |
|
|
3 |
|
Hospital's toilet |
Faucet handle |
|
|
|
1 |
2 |
1 |
4 |
|
|
Doorknob |
|
|
|
1 |
|
|
1 |
|
|
Toilet flush |
1 |
1 |
|
|
|
|
2 |
|
|
Toilet seat |
|
1 |
1 |
|
|
|
2 |
|
Public space |
Elevator button |
1 |
|
|
|
|
|
1 |
|
|
Keypad of ATM |
|
|
|
|
|
1 |
1 |
|
At University: |
|
|
|
|
|
|
|
|
|
Cafeteria's toilet |
Faucet handle |
|
|
|
|
1 |
|
1 |
|
|
Doorknob |
|
|
1 |
|
|
|
1 |
|
Gymnasium's toilet |
Faucet handle |
|
|
|
|
1 |
|
1 |
|
At petrol stations: |
|
|
|
|
|
|
|
|
|
Toilet |
Faucet handle |
|
|
|
|
1 |
|
1 |
|
Public space |
Keypad of ATM |
|
|
1 |
|
|
|
1 |
|
|
Total |
4 |
2 |
6 |
4 |
6 |
3 |
25 |
Note: Abbreviations: S. aureus, Staphylococcus aureus; CoNS, Coagulase-negative staphylococci; E. coli, Escherichia coli; K. pneumoniae, Klebsiella pneumoniae; P. aeruginosa, Pseudomonas aeruginosa; A. baumannii, Acinetobacter baumannii.
Alcohol susceptibility test
We hypothesized that a continuous exposure of bacteria with alcohol antiseptic during long term of COVID-19 pandemic, this stress may induce bacteria adaptation to stress response against alcohol decontamination. All 25 medically important bacteria were tested and compared the results of alcohol susceptibility test with bacteria collected before emerging of COVID-19 from ATCC strains and clinical isolates. As shown in Figure 1, all bacteria isolated in this study significantly exhibited a lower susceptibility to all types of alcohol tested when compared to ATCC strains and/or clinical isolates. Colonies within inhibition zone could be found in a comparative level between bacteria in this study and clinical strains.
Notably, Staphylococcus spp. isolated from environment in this study and clinical isolates were the most resistant bacteria due to presenting the narrowest inhibition zone with full of colonies growing inside (Figure 1 and 2). Comparison of antibacterial activity among three alcohol types, 75% IPA was better than 70% and 80% ethanol. For 70% ethanol preparation, additionally, two types of ethanol sources, denatured and absolute forms, were also compared to determine the efficacy of antibacteria by testing between ATCC strain and bacteria isolated in this study (Figure 3). It was found that 70% absolute ethanol had lesser or equal activity against bacteria when compared to 70% denatured ethanol
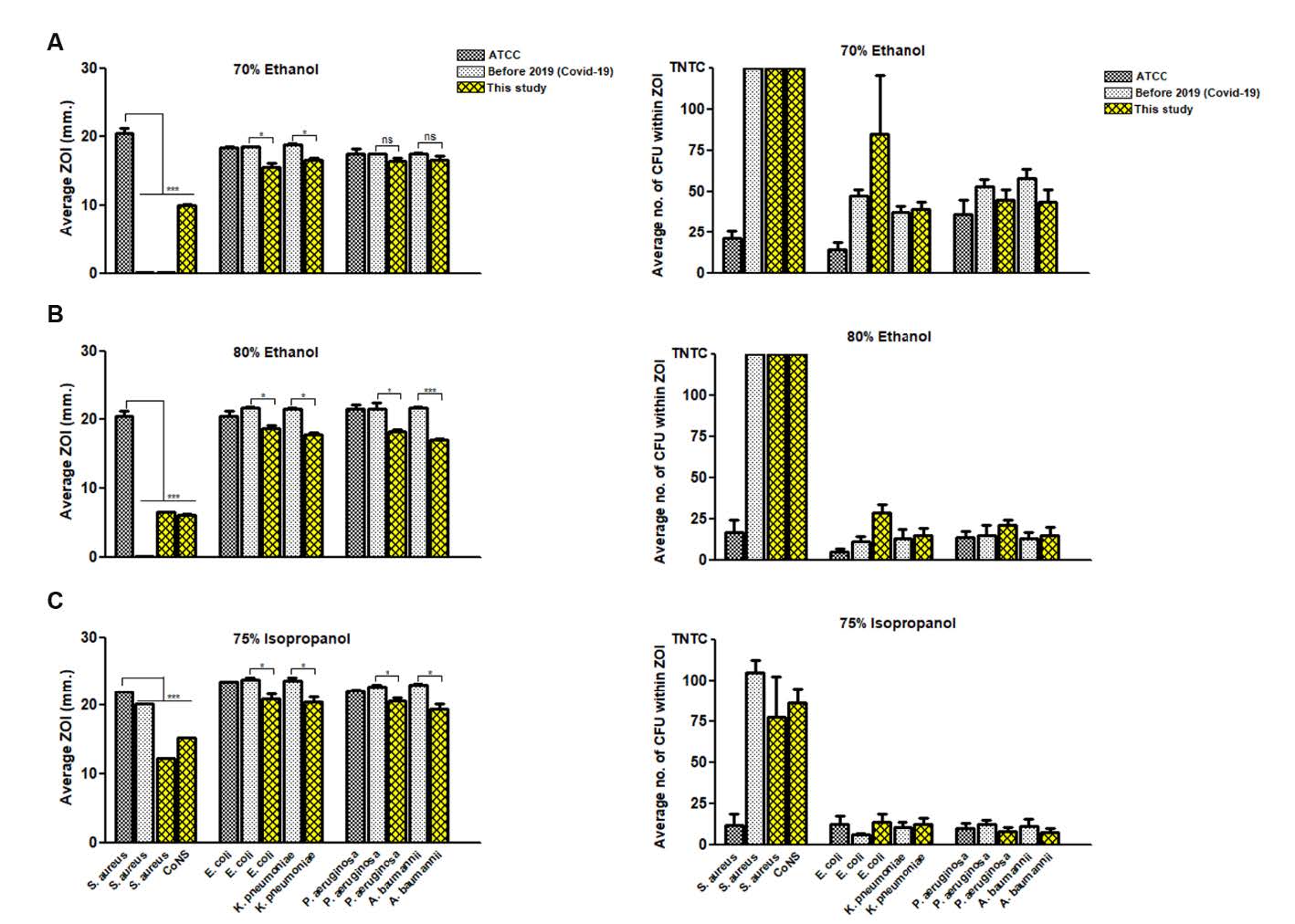
Figure 1. Alcohol susceptibility test. Tweny-five µl of (A) 70% ethanol, (B) 80% ethanol, and (C) 75% isopropanol was spotted on agar inoculated with each bacterial strain/isolate. After incubation period, (left panel) zone of inhibition (ZOI) was measured as millimeter (mm), and (right panel) the tolerance colonies growing within inhibition zone were counted as colony forming unit (CFU). The results of alcohol susceptibility test were compared among medically important bacteria isolated in this study (yellow bar), bacteria collected before COVID-19 pandemic from ATCC strains (dark grey bar) and clinical isolates (grey bar). Data are mean with SD from duplicate experiments. Statistical significance was determined with unpaired t-test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
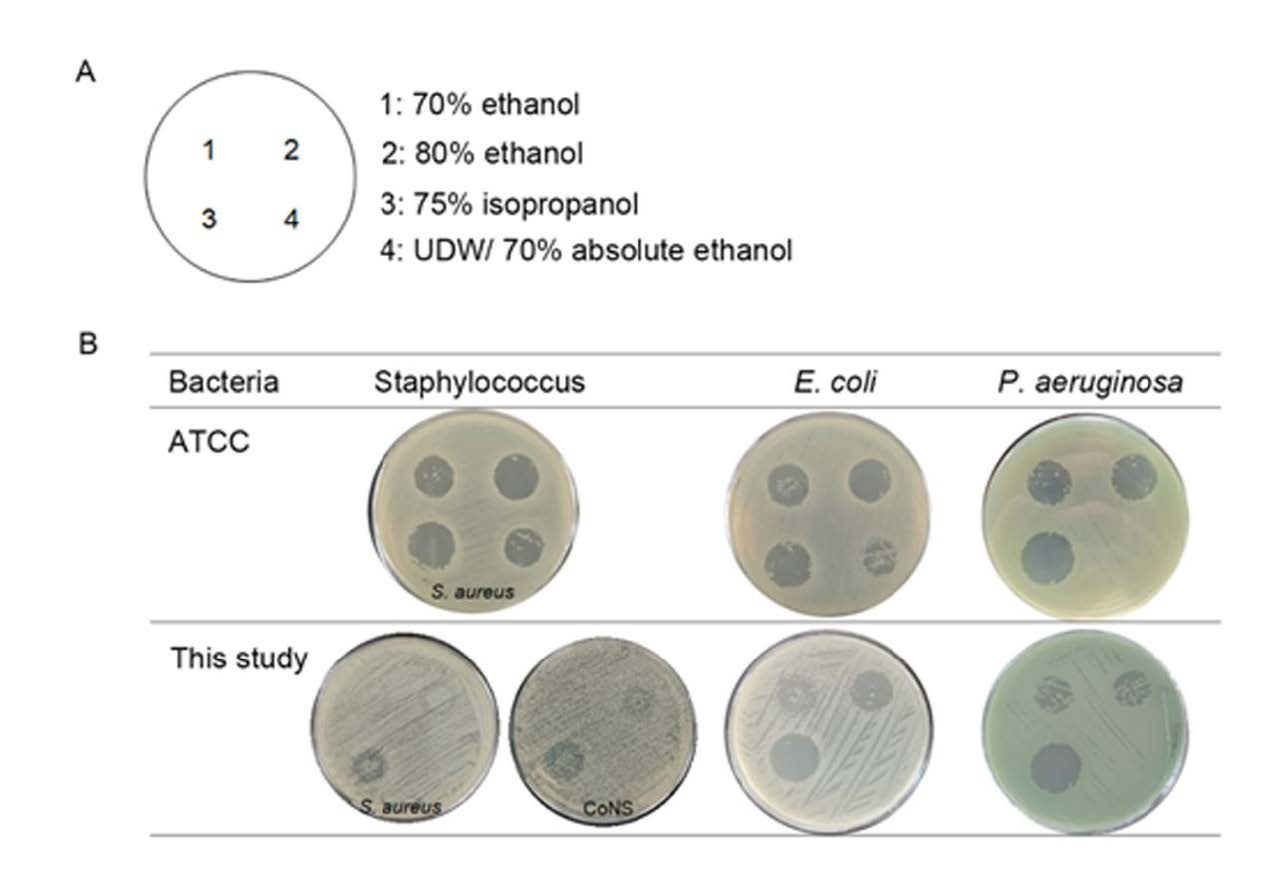
Figure 2. Illustration of inhibition zone by spot plating assay. (A) Different alcohol types and/or negative control (UDW) were spotted on MHA plate in the position as indicated. (B) Inhibition zones (mm) were compared among alcohol types and among bacteria from different sources. Here, inhibition zones between bacteria ATCC strain and isolates in this study were shown. Tolerant colonies growing within inhibition zone were also observed and counted.
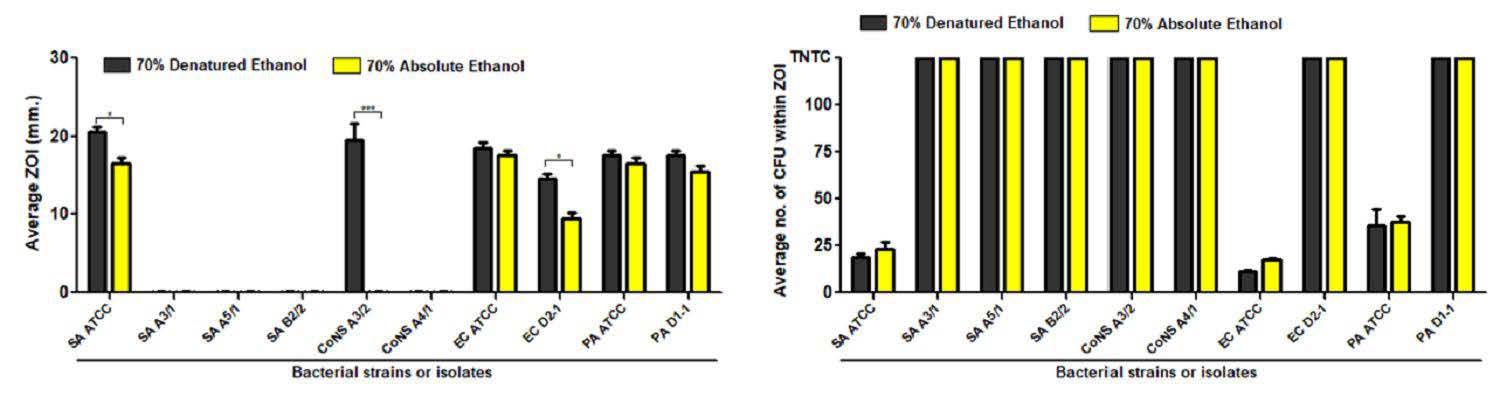
Figure 3. Comparison of antibacterial activity of 70% alcohol derived from different sources. Tweny-five µl of 70% denatured ethanol (black bar) and 70% absoute ethanol was spotted on agar inoculated with bacteria ATCC strain or isolate in this study. After incubation period, zone of inhibition (ZOI) was measured as millimeter (mm) (A), and the tolerance colonies growing within inhibition zone were counted as colony forming unit (CFU) (B). The results were compared between medically important bacteria isolated in this study and bacteria collected before COVID-19 pandemic from ATCC strains. Data are mean with SD from duplicate experiments. Statistical significance was determined with unpaired t-test. *, P < 0.05; ***, P < 0.001.
DISCUSSION
For SARS-CoV-2 detection in routine clinical laboratory, the Centers for Disease Control and Prevention (CDC) guideline only endorses using a synthetic fiber swab to collect nasopharyngeal sample (CDC, 2021). With limitation of high cost and shortage supply, it impeded to use a synthetic fiber swab for non-diagnostic purpose. Although some variations of testing can occur, our study indicated that using cotton swab can be applied to detect SARS-CoV-2 RNA from environmental samples. Thus, we recommended to use a cotton swab, inexpensive and readily available material, for a practical alternative procedure especially in developing countries.
In fact, general public places or even in hospitals that inhabited sites or quanrantine areas for COVID-19 patients are separately located from other parts have a low prevalence of SARS-CoV-2 contamination. A swab pooling assay, collecting swabs at the time of sample collection, was a suitable method to reduce cost of all required tests. Our study found contaminant SARS-CoV-2 in all samples at 8%. This seemed to be lower rate than other previous studies that the positive detection rate varied from 8.9% to more than 70% (Ben-Shmuel et al., 2020; Jiang et al., 2020; Lee et al., 2020; Dargahi et al., 2021; Moore et al., 2021; Wan et al., 2021). Factors contributing to the difference results involved with study sites, which most studies collected samples from COVID-19 isolation units, and infection rate among people in each study country.
Hospital’s toilets and frequently-touched surfaces were the higest risk area of SARS-CoV-2 contamination. This area should be concerned as an important hygiene site and must be always cleaned with effective disinfactants. Although average Ct value from positive samples in this study and others were in range of 34-38 (Ben-Shmuel et al., 2020; Jiang et al., 2020; Dargahi et al., 2021) and the limitation of molecular assays that cannot distinguish between infectious and non-infectious virus, we could not uncarefully conclude that the positive detections in environmental samples were from false positive result of remnant SARS-CoV-2 RNA or had a very low ability to transmit and infect hosts. Prolong persistence of infectious SARS-CoV-2 can be found on several types of materials in environments. The length of persistence involved with the suitable conditions of low temperature, high humidity, and metal material surface (Aboubakr et al., 2021).
Amid COVID-19 pandemic, alcohol-based hand sanitizers (ABHSs) are widely used as effective agent against both SARS-CoV-2 and bacteria. Two standard formulations of 80% v/v ethanol or 75% v/v isopropanol are recommended to use. However, the globally increasing demand for ABHSs to control the spreading of COVID-19 leads to abruptly shortage supply of standardized alcohol products. The number of substandard products are widely produced in the markets resulting in subinhibitory effect and induction of genetic mutations among resident bacteria on human hands after prolong exposure (Assefa and Melaku, 2021). The studies of bacterial gene expressions and omics technology revealed that several genes which are responsible for toxic or stress response pathways were associated with alcohol tolerant in bacteria (Cao et al., 2017; Horinouchi et al., 2018). Biofilm formation of B. cereus and Enterobacter cloacae environmental isolates associated with alcoholic tolerance by showing high survival rate up to 70% (Yeung et al., 2022). Thus, the genetic markers related to alcoholic resistance should be further investigated.
To our knowledge, our data provided the first evidence of a significant reduction in alcohol susceptibility test of all our bacterial isolates. However, 75% IPA was still the most effective agent against all groups of bacteria tested. This is due to the greater lipophilicity of isopropanol than other denatured ethanol (Singh et al., 2020). Therefore, the regulation of quality and appropriate use of ABHS should be effectively promoted.
CONCLUSION
In summary, this study emphasized that a prompt disinfection and cleaning of frequently touched surfaces in a high-risk area should be concerned as an effective infection control measure. Additionally, the proper use of ABHS is very important to prevent the global impacts of bacteria tolerance to alcohol after COVID-19 pandemic.
ACKNOWLEDGEMENTS
The RNA extraction reagent was supported by Thammasat University Research Unit in Modern Microbiology and Public Health Genomics, Thammasat University.
AUTHOR CONTRIBUTIONS
Nattamon Niyomdecha: Project administration, Conceptualization, Methodology, Investigation, Validation, Visualization, Formal Analysis, Writing- Original draft preparation. Pirom Noisumdaeng: Resources, Validation, Writing - Review & Editing. Pakkaporn Archawametheekul: Investigation. Sunisata Angkham: Investigation. Benja Norapong: Resources. Muttika Fungkrajai: Resources. Pramote Sriwanitchrak: Resources. All authors have read and approved of the final manuscript.
CONFLICT OF INTEREST
The authors declare that they hold no competing interests.
REFERENCES
Aboubakr, H.A., Sharafeldin, T.A., and Goyal, S.M. 2021. Stability of SARS-CoV-2 and other coronaviruses in the environment and on common touch surfaces and the influence of climatic conditions: A review. Transboundary and Emerging Diseases. 68: 296-312.
Assefa, D., and Melaku, T. 2021. Commercial hand sanitizers use amid COVID-19 pandemic: The concerns of antimicrobial resistance. Infection and Drug Resistance. 14: 2183-2185.
Ben-Shmuel, A., Brosh-Nissimov, T., Glinert, I., Bar-David, E., Sittner, A., Poni, R., and et al. 2020. Detection and infectivity potential of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) environmental contamination in isolation units and quarantine facilities. Clinical Microbiology and Infectious Diseases. 26: 1658-1662.
Cao, H., Wei, D., Yang, Y., Shang, Y., Li, G., Zhou, Y., and et al. 2017. Systems-level understanding of ethanol-induced stresses and adaptation in E. coli. Scientific Reports. 7: 1-15.
Castano, N., Cordts, S.C., Kurosu, Jalil. M., Zhang, K.S., Koppaka, S., Bick, A.D., and et al. 2021. Fomite transmission, physicochemical origin of virus-surface interactions, and disinfection strategies for enveloped viruses with applications to SARS-CoV-2. American Chemical Society. 6: 6509-6527.
Center for Diseases Control and Prevention [Internet]. 2021. Interim guidelines for collecting, handling, and testing clinical specimens from persons for Coronavirus Disease 2019 (COVID-19). [Updated 2021 Oct 25; Cited 2021 Dec 15]. Available from: https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specimens.html.
Cervino, G.F., Surace, L., Paduano, G., Fiorillo, V., De Stefano, M.T., Laudicella, R., and et al. 2020. SARS-CoV-2 persistence: Data summary up to Q2 2020. Data. 5: 81.
Chojnacki, M., Dobrotka, C., Osborn, R., Johnson, W., Young, M., Meyer, B., and et al. 2021. Evaluating the antimicrobial properties of commercial hand sanitizers. mSphere. 6: e00062-21.
Corman, V.M., Landt, O., Kaiser, M., Molenkamp, R., Meijer, A., Chu, D.K., and et al. 2020. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 25.
Dargahi, A., Jeddi, F., Vosoughi, M., Karami, C., Hadisi, A., Ahamad., Mokhtari, S., and et al. 2021. Investigation of SARS CoV-2 virus in environmental surface. Environmental Research. 195: 110765.
Food and Drug Administration [Internet]. 2020. Policy for temporary compounding of certain alcohol-based hand sanitizer products during the public health emergency immediately in effect: guidance for industry. [Cited 2021 Dec 15]. Available from: https://www.fda.gov/drugs/guidance-compliance-regulatory-information/guidances-drugs.
Horinouchi, T., Maeda, T., and Furusawa, C. 2018. Understanding and engineering alcohol-tolerant bacteria using OMICS technology. World Journal of Microbiology and Biotechnology. 34: 1-9.
Jiang, F.C., Jiang, X.L., Wang, Z.G., Meng, Z.H., Shao, S.F., Anderson, B.D., and et al. 2020. Detection of severe acute respiratory syndrome coronavirus 2 RNA on surfaces in quarantine rooms. Emerging Infectious Diseases. 26: 2162–2164.
Lee, S.E., Lee, D.Y., Lee, W.G., Kang, B., Jang, Y.S., Ryu, B., and et al. 2020. Detection of novel coronavirus on the surface of environmental materials contaminated by COVID-19 patients in the Republic of Korea. Osong Public Health and Research Perspectives. 11: 128-32.
Mahmood, A., Eqan, M., Pervez, S., Alghamdi, H.A., Tabinda, A.B., Yasar, A., and et al. 2020. COVID-19 and frequent use of hand sanitizers; human health and environmental hazards by exposure pathways. The Science of the Total Environment. 742: 140561.
Mohamadi, M., Babington-Ashaye, A., Lefort, A., and Flahault, A. 2021. Risks of infection with SARS-CoV-2 due to contaminated surfaces: A scoping review. International Journal of Environmental Research and Public Health. 18.
Moore, G., Rickard, H., Stevenson, D., Aranega-Bou, P., Pitman, J., Crook, A., and et al. 2021. Detection of SARS-CoV-2 within the healthcare environment: a multi-centre study conducted during the first wave of the COVID-19 outbreak in England. The Journal of Hospital Infection.108: 189-96.
Pastorino, B., Touret, F., Gilles, M., de Lamballerie, X., and Charrel, R.N. 2020. Prolonged infectivity of SARS-CoV-2 in fomites. Emerging Infectious Diseases. 26: 2256-2257.
Pidot, S.J., Gao, W., Buultjens, A.H., Monk, I.R., Guerillot, R., Carter, G.P., and et al. 2018. Increasing tolerance of hospital Enterococcus faecium to handwash alcohols. Science Translational Medicine. 10: eaar6115.
Singh, D., Joshi, K., Samuel, A., Patra, J., and Mahindroo, N. 2020. Alcohol-based hand sanitisers as first line of defence against SARS-CoV-2: A review of biology, chemistry and formulations. Epidemiology and Infection.148: e229.
Wan, B., Zhang, X., Luo, D., Zhang, T., Chen, X., Yao, Y., and et al. 2021. On-site analysis of COVID-19 on the surfaces in wards. The Science of the Total Environment. 753: 141758.
Yeung, Y.W.S., Ma, Y., Liu, S.Y., Pun, W.H., and Chua, S.L. 2022. Prevalence of alcohol-tolerant and antibiotic-resistant bacterial pathogens on public hand sanitizer dispensers. Journal of Hospital Infection. 127: 26-33.
OPEN access freely available online
Natural and Life Sciences Communications
Chiang Mai University, Thailand.
Nattamon Niyomdecha1,*, Pirom Noisumdaeng2, 3, Pakkaporn Archawametheekul1, Sunisata Angkham1, Benja Norapong1, Muttika Fungkrajai1, and Pramote Sriwanitchrak1
1 Department of Medical Technology, Faculty of Allied Health Sciences, Thammasat University, Rangsit Campus, Pathum Thani 12121, Thailand
2 Faculty of Public Health, Thammasat University, Rangsit Campus, Pathum Thani 12121, Thailand
3 Thammasat University Research Unit in Modern Microbiology and Public Health Genomics, Thammasat University, Pathum Thani 12121, Thailand.
Corresponding author: Nattamon Niyomdecha, E-mail: nattamon@tu.ac.th
Total Article Views
Editor: Wasu Pathom-aree,
Chiang Mai University, Thailand
Article history:
Received: May 5, 2022;
Revised: October 29, 2022;
Accepted: November 4, 2022;
Published online: November 16, 2022

