
The Anti-Inflammatory Potential of Red Betel (Piper crocatum) Leaves Through Inhibitory Mechanism on Nfκb Signaling Pathway: Drug-Like Candidate Study
Siti Imroatul Maslikah*, Sri Rahayu Lestari, Nursasi Handayani, Wira Eka Putra, Alif Rofiqotun Nurul Alimah, Atikah Amalia, Solichatul Afifah, and Siti Nur ArifahPublished Date : 2023-01-05
DOI : https://doi.org/10.12982/NLSC.2023.005
Journal Issues : Number 1, January-March 2023
Abstract Rheumatoid arthritis (RA) is a systemic autoimmune disease that causes inflammation of the synovial tissue bone as well as joint damage. Furthermore, an increase in the level of pro-inflammatory cytokines, such as tumor necrosis factor α (TNF-α) due to overexpression in the nuclear factor-kappa B (NFκB) contributes to the progression of the disease. NFκB also plays an important role in the production, differentiation, and effector function of inflammatory T cells. Red betel (Piper crocatum) leaf (RBL) is an Indonesian herb, which contains bioactive compounds such as flavonoids, terpenoids, and phenolic compounds. It is widely used as an intervention for various diseases including inflammatory-related diseases. Therefore, this study aims to evaluate the therapeutic effect of RBL extract as an anti-inflammatory agent through inhibition on the TNF receptor 1(TNFR1), NFκB, and inhibitor kappa B kinase (IκK) by molecular docking study. Oral toxicity prediction was carried out before molecular docking. Molecular docking performed using PyRx 0.8 software. The amino acid residues analysis and visualization were conducted using the Biovia Discovery Studio and Pymol. The toxicity prediction using ProTox-II showed that RBL active compounds are categorized between the 4th-6th class. Furthermore, the compounds, specifically kaempferitrin and apigenin have greater binding affinity compared to the drug inhibitor in NFκB signalling pathway. Based on the results, RBL active compounds can potentially act as an anti-inflammatory agent in RA, but further studies must be carried out to explore the potency of RBL through in vitro and in vivo effects.
Keywords: Red betel leaf, Molecular docking, Rheumatoid arthritis, Flavonoid
Citation: Maslikah, S.I., Lestari, S.R., Handayani, N., Putra, W.E., Alimah, A.R.N., Amalia, A., Afifah, S., and Arifah, S.N. 2023. The anti-inflammatory potential of red betel (Piper crocatum) leaves through inhibitory mechanism on NFΚB signaling pathway: Drug-like candidate study. Nat. Life Sci. Commun. 22(1): e2023005.
INTRODUCTION
Rheumatoid arthritis (RA) is a chronic disease characterized by inflammation of the synovial tissue, which causes bone and cartilage damage (Tsubaki et al., 2015). Furthermore, it reduces the patients' quality of life because their movements are limited (Almoallim et al., 2021). Several studies reported that the cause of RA is still unclear, but genetic abnormalities (autoimmune), age, and environmental factors, such as socioeconomic status and ethnicity can trigger inflammatory conditions (Okada et al., 2014). For some cases with RA in family members could increase the coincidence of RA up to 40-60% (Smolen et al., 2016). In 2017, the Global Burden of Disease, Injuries, and Risk Factor (GBD) stated that there are approximately 20 million RA patients in the world (Safiri et al., 2019). Furthermore, it is more prevalent in women than men with a ratio of 3:1 (Intriago et al., 2019) due to hormonal factors (Safiri et al., 2019; Almoallim et al., 2021).
RA has been reported to be associated with the up-regulation of inflammatory cytokines or other inflammatory mediators. Furthermore, several pro-inflammatory cytokines are involved in its progression including tumor necrosis factor (TNF)-α, interleukin (IL)-1, IL-6, and IL17 (Alghasham & Rasheed, 2014; Alunno et al., 2017). These mediators are produced through a downstream pathway, which causes damage to the cartilage and bone (Alunno et al., 2017). The nuclear factor kappa B (NFκB) is a member of the transcription factor group, which is responsible for the expression of many genes that are involved in the inflammatory mechanism and immunity (MacArthur et al., 2017). The NFκB family plays a crucial role in the activation, differentiation, and effector function of inflammatory T cells. They also consist of NFκB1 (p50), NFκB2 (p52), RelA (p65), RelB, and c-Rel, and are involved in gene transcription by forming different types of heterodimers (T. Liu et al., 2017). NFκB proteins are often inactive due to the presence of inhibitory proteins family, namely inhibitory kappa B (IκB). However, they are activated by several intermediate mechanisms, which lead to their interaction with IκB kinase (IκK). This activation causes the expression of RA, hence, it is very essential to regulate NFkB (T. Liu et al., 2017).
For decades, betel leaves have been widely used as herbal therapy. The plant was first discovered in India, but it has gained popularity amongst other South Asian countries, including Indonesia (Gundala & Aneja, 2014). Furthermore, its leaves are traditionally used to treat several conditions, such as itches, abscesses, constipation, abrasions, and rheumatism. Previous studies reported that the therapeutic effect of betel can be attributed to the antioxidant, anti-microbial, antifungal, antimutagenic, and chemo-preventive activities of its constituents (Sarma et al., 2018). In Indonesia, there are various varieties of the plant, such as green betel (Piper betel) and red betel (Piper crocatum). Red betel is characterized by its red color in branching vines and silver-red heart-shaped leaves. It contains flavonoids, tannins, alkaloids, saponins, steroids, and essential oil (Gupta et al., 2012). Its other constituents include phenolic compounds, such as hydroxychavicol, eugenol, chavibetol, piperbetol, and piperine, which were known as antimutagenic agents (Chang et al., 2002; Gundala & Aneja, 2014). Due to the high phenolic and flavonoid content of red betel, it is very important to explore the role of its leaves in the inflammatory pathway. Therefore, this study aims to elucidate the mechanism of red betel leaves’ active compound in the TNF activation pathway.
MATERIALS AND METHODS
Ligand Preparation
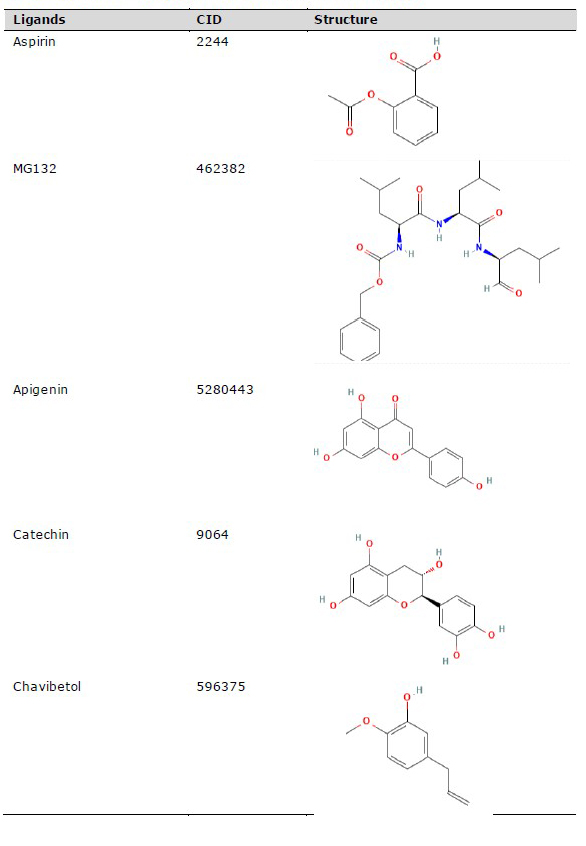
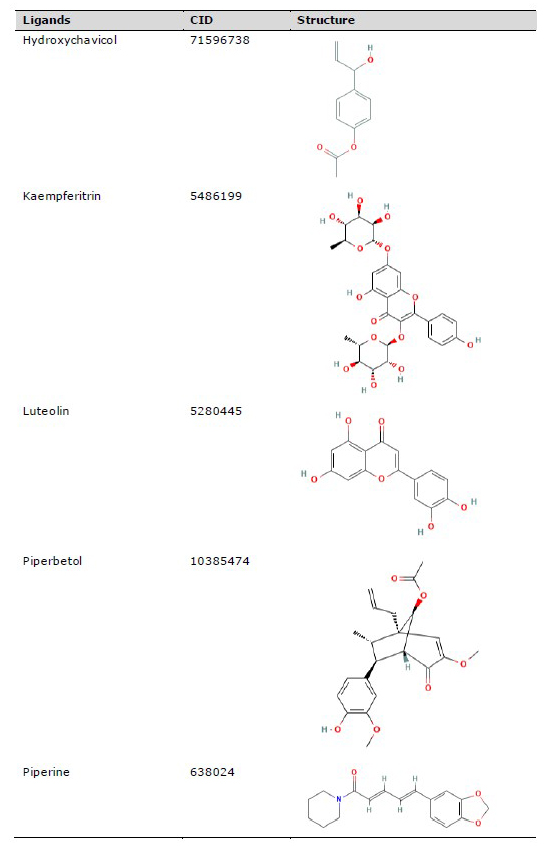
The three-dimensional (3D) structure of red betel active compounds was obtained from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/), as shown in Table 1. The structure was saved in .sdf format and then converted into .pdb format using the PyMOL software. Aspirin was used as the drug inhibitor of TNRF1, while MG132 (N-carbobenzonyl-Leu-Leu-leucinal) served as the inhibitor for NFκB p52/RelB complex and IκK (Kadioglu et al., 2015).
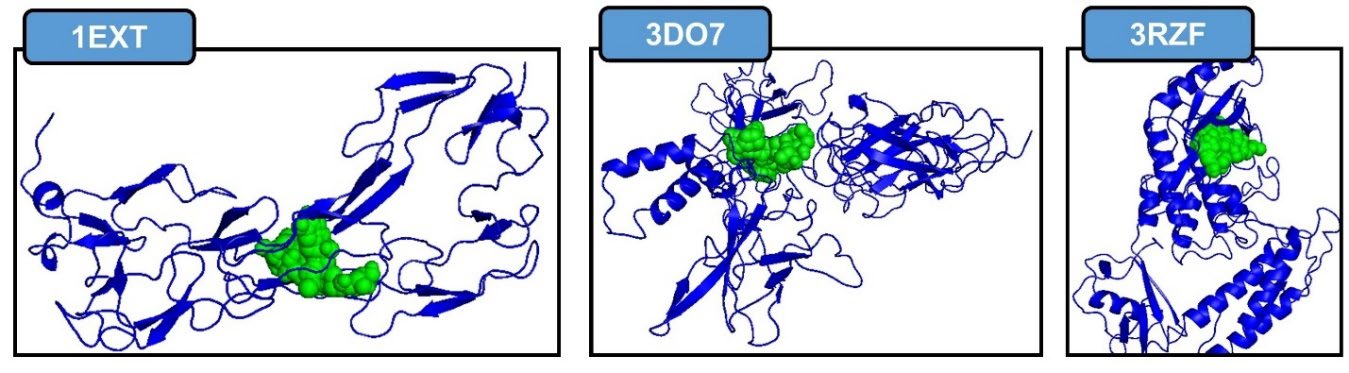
Figure 1. Active site for each protein. TNFR1, NFκB p52/RelB/DNA complex, IκK. Blue color as protein, green color as ligand.
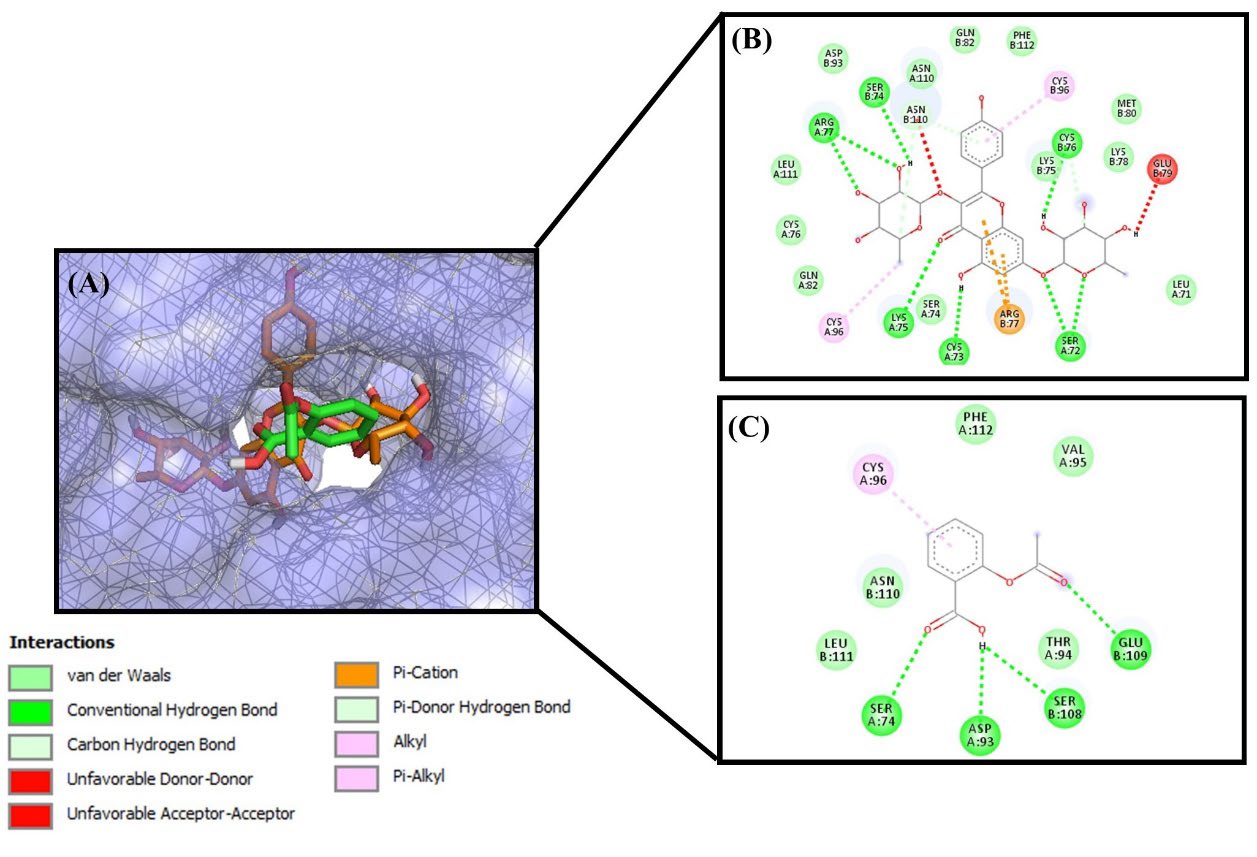
Figure 2. Molecular docking between ligand and TNFR1 protein. (A) Active site of TNFR1 with kaempferitrin (orange) and aspirin (green) as ligand. 2D interaction and amino acid residues (B) kaempferitrin and (C) aspirin.
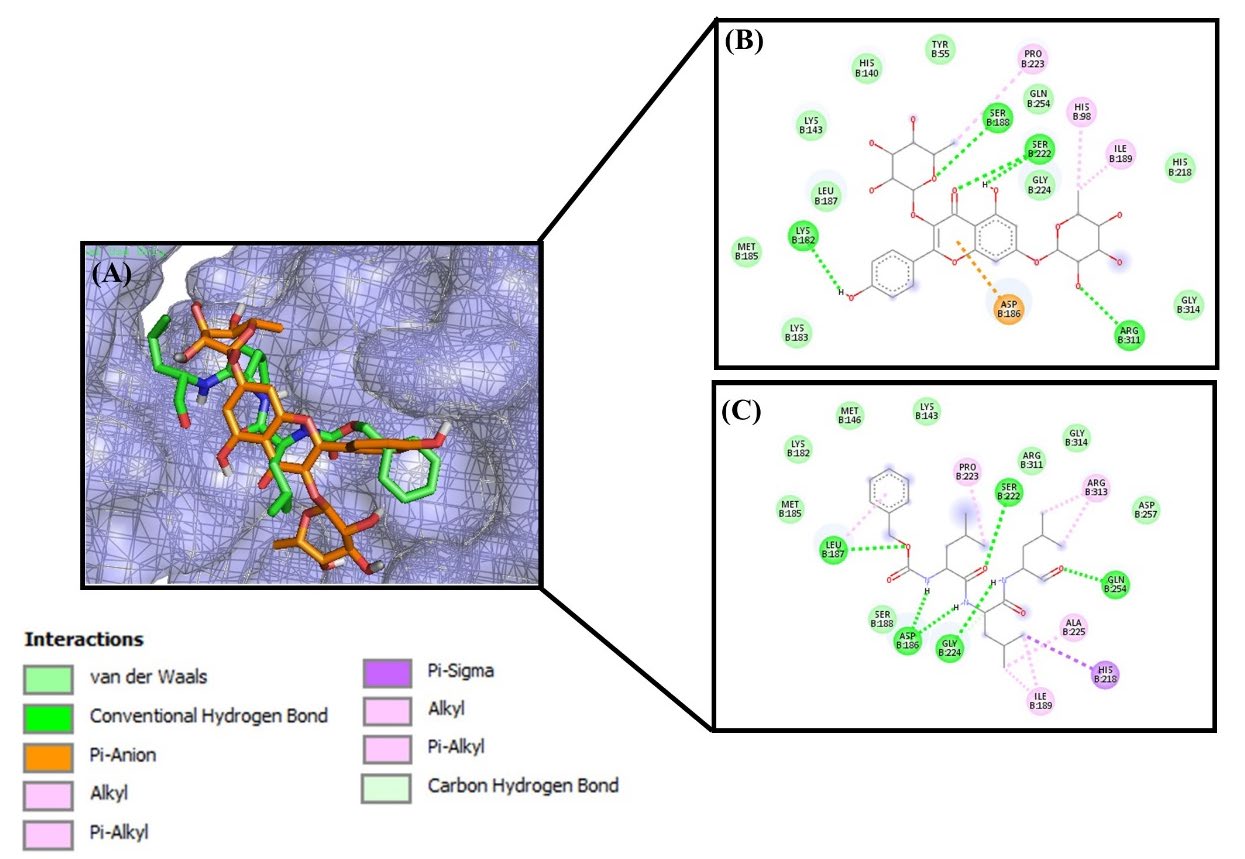
Figure 3. Molecular docking between ligand and NFκB p52/RelB/DNA protein complex. (A) Active cite of p52/RelB/DNA protein complex with kaempferitrin (orange) and MG132 (green) as ligand. 2D interaction and amino acid residues (B) kaempferitrin and (C) MG132

Figure 4. Molecular docking between ligand and IκK protein. (A) Active site of IκK protein with apigenin (white) and MG132 (green). 2D interaction and amino acid residues (B) apigenin and (C) MG132.
Table 1. Chemical compounds for molecular docking study.
Oral Toxicity Prediction
Oral toxicity prediction was carried out with the ProTox-II (https://tox-new.charite.de) online webserver. It is essential to perform a prediction before drug design because it helps to validate whether the ligand is safe for consumption. The lethal doses 50 (LD50), hepatoxicity, carcinogenicity, immunotoxicity, mutagenicity, and cytotoxicity of the ligands were also included in the prediction (Drwal et al., 2014; Banerjee, Dehnbostel, et al., 2018; Banerjee, Eckert, et al., 2018). Oral toxicity predictions were classified into 6 classes, namely:
Class I is fatal when swallowed (LD50≤5)
Class II is fatal when swallowed (5
Class III is toxic when swallowed (50
Class IV is harmful when swallowed (300
Class V is potentially harmful when swallowed (2000
Class VI is non-toxic (LD50>5,000)
Pharmacokinetics Prediction
The pharmacokinetics prediction was examined using pkCSM online tool (http://biosig.unimelb.edu.au/pkcsm/prediction). Prediction of pharmacokinetic included absorption, distribution, metabolism, excretion, and toxicity (ADMET). The parameters that were examined in absorption included Caco2 permeability and intestinal absorption in human, while distribution study was determined by volume of distribution (VDss) and blood-brain barrier (BBB) in human. For metabolism study, the prediction was analyzed including cytochrome P450 Family 2 Subfamily D Member 6 or CYP2D6 substrate, CYP3A4 substrate, CYP1A2 inhibitor, CYP2C19 inhibitor, CYP2C9 inhibitor, CYP2D6 inhibitor, and CYP3A4 inhibitor. The pharmacokinetic properties for excretion included total clearance and renal organic cation transporter 2 (OCT2) substrate. Furthermore, for toxicity test were using Ames toxicity.
Protein Preparation
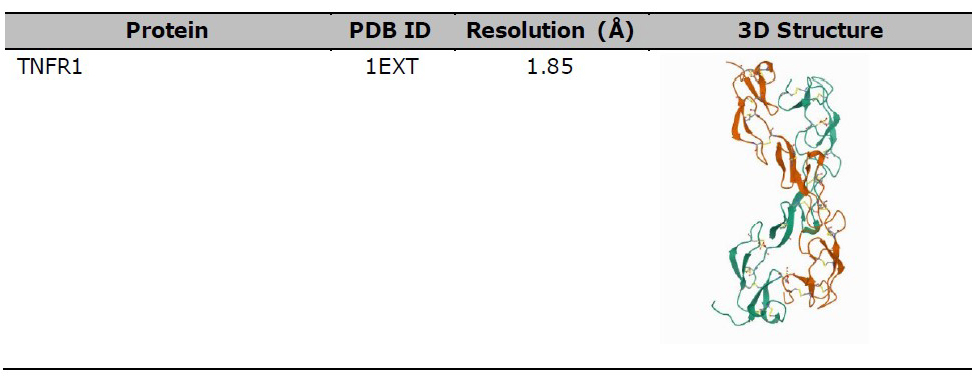
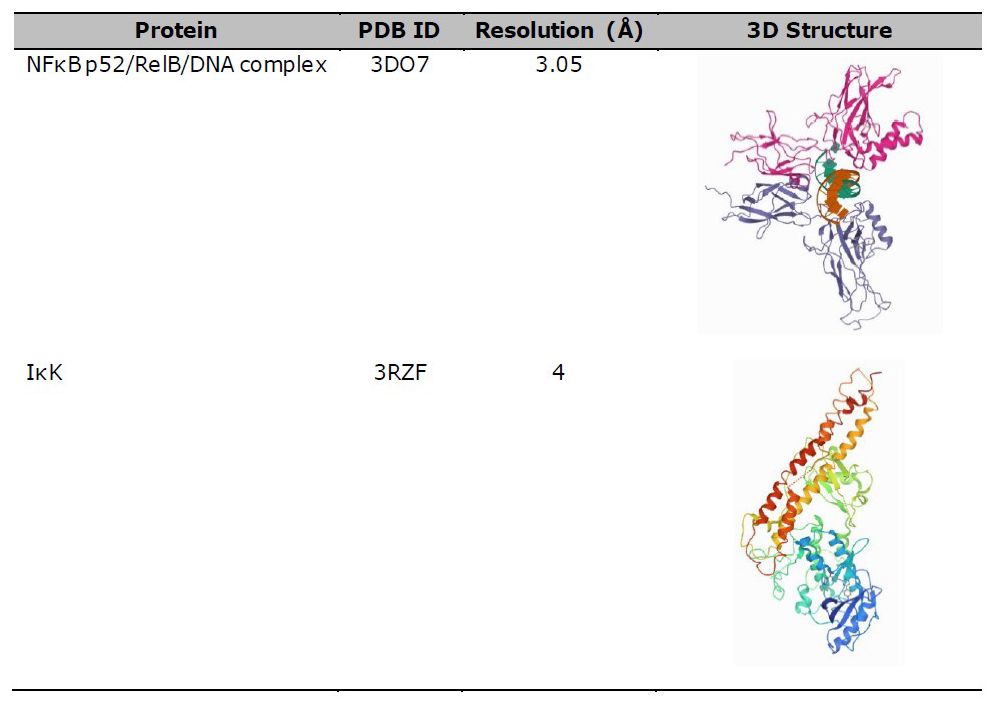
TNFR1 (PDB 1EXT), NFκB (PDB 3DO7), and IκK (PDB 3RZF) were used as protein targets for the molecular docking study between protein-ligand, as shown in Table 2. The protein structure was downloaded from the Protein Data Bank (https/www.rscb.org/) in .pdb format, after which water and ligands were removed using Pymol software version 1.7.4.5 Edu (Schrӧdinger Inc., LLC). TNFR1 was then selected as the ligand-protein and compared with the result of protein-protein docking by Chen et al. (2017). Meanwhile, NFκB and IκK were used as the protein target based on the steps outlined by Kadioglu et al. (2015).
Table 2. 3D structure of protein for molecular docking study.
Molecular Docking Simulation
Molecular docking analysis was carried out using Autodock Vina and Pyrx v.0.8 (https/pyrx.sourceforge.io) (Dallakyan & Olson, 2015; Trott & Olson, 2010). The ligands were minimized using the Open Babel GUI to reduce their energy, consequently, the protein-ligand interactions were similar to the quantum chemical calculations levels (Mirzaei et al., 2015). Table 3 shows the grid center of docking for each protein. The binding affinity values of each ligand were analyzed, and the values were indicated by their bonding strength with protein. Furthermore, 2D interaction of amino acid residues was visualized using the BIOVIA Discovery Studio (Dassault Systèmes BIOVIA, 2015), while visualization of protein-ligand site was performed with the PyMol software version 1.7.4.5 Edu (Schrӧdinger Inc., LLC).
Table 3. Grid center for each protein of molecular docking study.
|
Protein |
Grid center |
||
|
x |
y |
z |
|
|
TNFR1 |
3.949 |
37.693 |
-9.44 |
|
NFκB p52/RelB/DNA complex |
26.376 |
60.42 |
75.564 |
|
IκK |
88.492 |
-29.695 |
56.299 |
Protein Network Interaction
Analysis of protein network interaction in NFκB and TNF-mediated signaling pathways was carried out using the Search Tool for the Retrieval of Interacting Genes/Protein (STRING) database online webserver (http://string-db.org/) (Szklarczyk et al., 2015). This was performed to measure the proteins that are involved in certain pathways. Subsequently, ligand-protein network interaction analysis was carried out with the STITCH database online webserver (www.stitch.embl.de). The web server allows the navigation and visualization of networks between chemicals as well as their interaction with proteins (Kuhn et al., 2008).
RESULTS
Oral Toxicity Prediction
The oral toxicity prediction result of RBL active compounds was categorized into class IV-VI. Chavibetol, hydroxychavicol, piperbetol, and piperine were in class IV, while kaempferitrin and luteolin were in Class V, and catechin was in class VI, as shown in Table 4. Meanwhile, drug inhibitors namely aspirin and MG1132 were in class III and class V, respectively. These results indicate that RBL is not toxic when swallowed (under the LD50) and it can be used for drug development. The prediction results also showed that aspirin was toxic when swallowed at LD50 of 250 mg/kg.
Table 4. Oral toxicity prediction results.
|
Compounds |
LD50 (mg/kg) |
Class |
Hpt |
Crg |
Imm |
Mtgn |
Ctcy |
|
Aspirin |
250 |
III |
(-) |
(-) |
(-) |
(-) |
(-) |
|
MG132 |
2.025 |
V |
(-) |
(-) |
(-) |
(-) |
(-) |
|
Apigenin |
2.500 |
V |
(-) |
(-) |
(-) |
(-) |
(-) |
|
Catechin |
10.000 |
VI |
(-) |
(-) |
(-) |
(-) |
(-) |
|
Chavibetol |
1.230 |
IV |
(-) |
(-) |
(-) |
(-) |
(-) |
|
Hydroxychavicol |
1.930 |
IV |
(-) |
(+) |
(-) |
(+) |
(-) |
|
Kaempferitrin |
5.000 |
V |
(-) |
(-) |
(+) |
(-) |
(-) |
|
Luteolin |
3.919 |
V |
(-) |
(+) |
(-) |
(+) |
(-) |
|
Piperbetol |
1.050 |
IV |
(-) |
(-) |
(+) |
(-) |
(-) |
|
Piperine |
330 |
IV |
(-) |
(+) |
(+) |
(-) |
(-) |
Note: Hepatoxicity, Hpt; Carcinogenicity, Crg; Immunotoxicity, Imm; Mutagenicity, Mtg; Cytotoxicity, Ctcy; active (+); inactive (-)
Pharmacokinetics Prediction
Based on the results of pharmacokinetics properties prediction. The absorption of all compounds include aspirin, MG132, and RBL active compounds showed that piperin has the highest absorption in permeability of Caco-2 cells. In the other hands, piperbetol, piperin, apigenin, hydroxychavicol, and chavibetol reached the absorption in the human intestinal up to 90% (Table 5). Based on distribution volume (VDss) analysis showed that all compounds has relatively low VDss (Han
et al., 2019). BBB analysis showed that chavibetol and hydrovychavicol have logBB > 0.3 and this is indicated that those compounds could easily cross the blood-brain barrier.
Cytochrome P450s (CYP) is an important enzyme system for metabolism the drug in the liver. Metabolism analysis suggested that catechin, hydroxychavicol, and kaempferitrin are not metabolized in the liver. In the excretion prediction, the renal total clearance for RBL active compounds have a good excretion. Renal OCT2 analysis showed that RBL active compound could eliminate completely such as apigenin, catechin, hydroxychavicol, kaempferitrin, luteolin, and piperbetol. For toxicity test using Ames showed that apigenin, catechin, hydroxychavicol, kaempferitrin, luteolin, piperbetol, and piperin has no mutagenic and carcinogenic agent.
Table 5. Pharmacokinetics study of control drug and RBL active compounds.
|
Parameters |
Compounds |
||||||||||
|
Aspirin |
MG132 |
Apigenin |
Catechin |
Chavibetol |
Hydroxychavicol |
Kaempferitrin |
Luteolin |
Piperbetol |
Piperin |
||
|
Absorption |
Caco-2 permeability (log Papp in 10-6 cm/s) |
0.09 |
0.77 |
1.007 |
-0.283 |
1.497 |
1.676 |
0.225 |
0.096 |
1.301 |
1.596 |
|
Intestinal absorption (human) (% Absorbed) |
76.938 |
64.418 |
93.25 |
68.829 |
91.835 |
92.09 |
35.385 |
81.13 |
96.282 |
94.444 |
|
|
Distribution |
VDss (human) |
-1.716 |
0.424 |
0.822 |
1.027 |
0.203 |
0.477 |
1.487 |
1.153 |
0.107 |
0.158 |
|
BBB permeability |
-0.332 |
-0.955 |
-0.734 |
-1.054 |
0.389 |
0.361 |
-1.823 |
-0.907 |
-0.522 |
-0.102 |
|
|
Metabolism |
CYP2D6 substrate |
No |
No |
No |
No |
No |
No |
No |
No |
No |
No |
|
CYP3A4 substrate |
No |
Yes |
No |
No |
No |
No |
No |
No |
Yes |
Yes |
|
|
CYP1A2 inhibitor |
No |
No |
Yes |
No |
Yes |
No |
No |
Yes |
No |
No |
|
|
CYP2C19 inhibitor |
No |
Yes |
Yes |
No |
No |
No |
No |
No |
No |
Yes |
|
|
CYP2C9 inhibitor |
No |
No |
No |
No |
No |
No |
No |
Yes |
No |
No |
|
|
CYP2D6 inhibitor |
No |
No |
No |
No |
No |
No |
No |
No |
No |
No |
|
|
CYP3A4 inhibitor |
No |
Yes |
No |
No |
No |
No |
No |
No |
No |
No |
|
|
Excretion |
Total Clearance |
0.72 |
1.14 |
0.566 |
0.183 |
0.28 |
0.206 |
-0.102 |
0.495 |
0.295 |
0.232 |
|
Renal OCT2 substrate |
No |
No |
No |
No |
No |
No |
No |
No |
No |
Yes |
|
|
Toxicity |
AMES toxicity |
No |
No |
No |
No |
Yes |
No |
No |
No |
No |
No |
Molecular Docking Study
The computation study of molecular docking was based on the binding affinity value between the ligand and target protein complex. The results revealed that RBL active compounds have a higher bond affinity for TNFR1 compared to aspirin as a drug inhibitor, as shown in Table 6. Based on the results, the binding affinity value for the aspirin-TNFR1 complex was -5.8 kcal/mol. This value was higher compared to other RBL active compounds, such as apigenin, catechin, kaempferitrin, luteolin, piperbetol, and piperin with binding affinity values of -7.7, -8.4, -9.8, -8.1, -8.0, and -7.5 respectively, thus the binding affinity of asipirin is weaker than those compounds. Furthermore, these results indicated that kaempferitrin had the strongest binding affinity compared to other active constituents.
MG132 served as a drug inhibitor for NFκB p52/RelB/DNA complex and IκK (Kadioglu et al., 2015). The molecular docking result showed that its binding affinity value with NFκB p52/RelB/DNA complex and IκK was -6.8 and -7.2, respectively, as shown in Table 6. In the NFκB p52/RelB/DNA protein complex study, kaempferitrin was stronger than MG132 and also became the strongest compound to bind with it. Meanwhile, in the IκK protein study, there were several RBL compounds, with stronger bond compared to MG132, namely apigenin, catechin, kaempferitrin, luteolin, piperbetol, and piperin. They had a binding affinity of -8.3, -7.9, -8.3, -8.2, -7.5, and -8.1, respectively, as shown in Table 6. Furthermore, apigenin had the highest value compared to the other ligands, and these results indicate that RBL active compounds such as kaempferitrin, apigenin, and catechin are better than anti-inflammatory drugs.
Table 6. Binding affinity value from molecular docking study.
|
Ligands |
Binding Affinity Value (kcal/mol) |
|||
|
TNFR1 (1EXT) |
NFκB p52/RelB/DNA complex(3DO7) |
IκK (3RZF) |
||
|
Aspirin (drug inhibitor for 1EXT) |
-5.8 |
- |
- |
|
|
MG132 (drug inhibitor for 3DO7 and 3RZF) |
- |
-6.8 |
-7.2 |
|
|
Red betel compounds |
|
|
|
|
|
|
Apigenin |
-7.7 |
-6.2 |
-8.3 |
|
|
Catechin |
-8.2 |
-6.1 |
-7.9 |
|
|
Chavibetol |
-5.5 |
-4.4 |
-5.5 |
|
|
Hydroxychavicol |
-5.4 |
-4.6 |
-5.6 |
|
|
Kaempferitrin |
-9.8 |
-7.6 |
-8.2 |
|
|
Luteolin |
-8.1 |
-6.5 |
-8.2 |
|
|
Piperbetol |
-8.0 |
-6.1 |
-7.5 |
|
|
Piperine |
-7.5 |
-6.3 |
-8.1 |
Protein Interaction
The protein network interaction with the STRING database was used to analyze proteins that are involved in the inflammatory pathways, namely including NFκ.B signaling and TNF-mediated signaling pathways. The STRING analysis result showed that there was an interaction between both signaling pathways. Furthermore, several proteins were involved including TNF, TNFR Sub Family 1A (TNFRSF1A), TNF Receptor Associated Factor 2 (TRAF2), TNF receptor type 1-associated death domain (TRADD), receptor-interacting serine/threonine-protein kinase (RIPK1), conversed helix-loop-helix ubiquitous kinase (CHUK)/ IκK-α, IκK subunit beta (IKBKB), RELA, and NFκB Inhibitor α (NFKBIA), as shown in Figure 5.
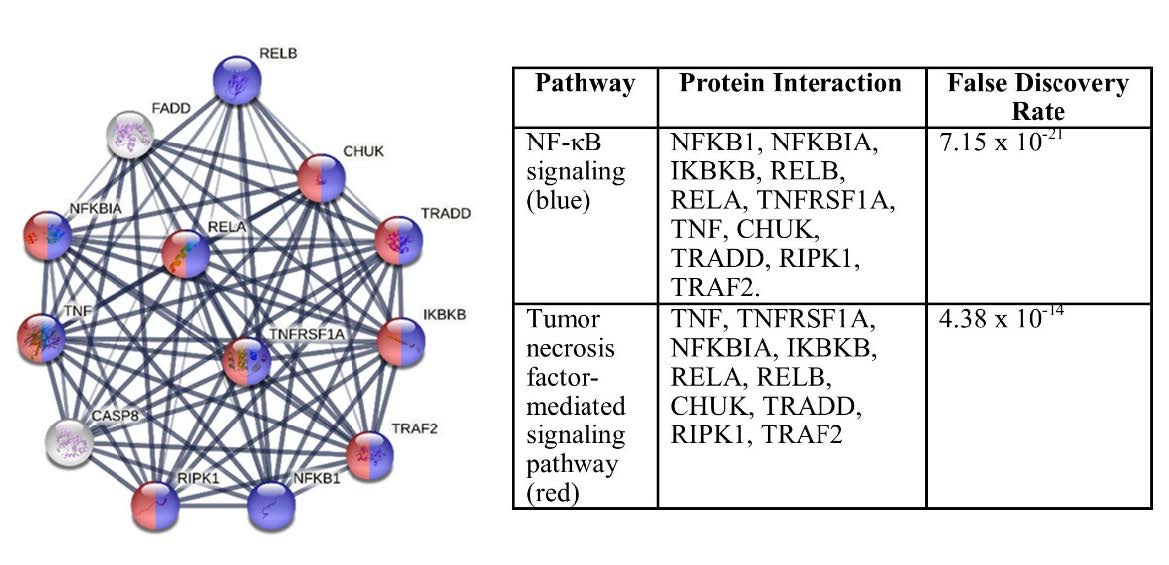
Figure 5. STRING analysis of protein network interactions.
Based on STITCH analysis, RBL active compounds, namely catechin, apigenin, luteolin, and piperine had direct bonding interaction with the proteins involved in the inflammation process (red ball), as shown in Figure 6. However, the interaction of chavibetol, hydroxychavicol, and kaempferitrin is still unreported.
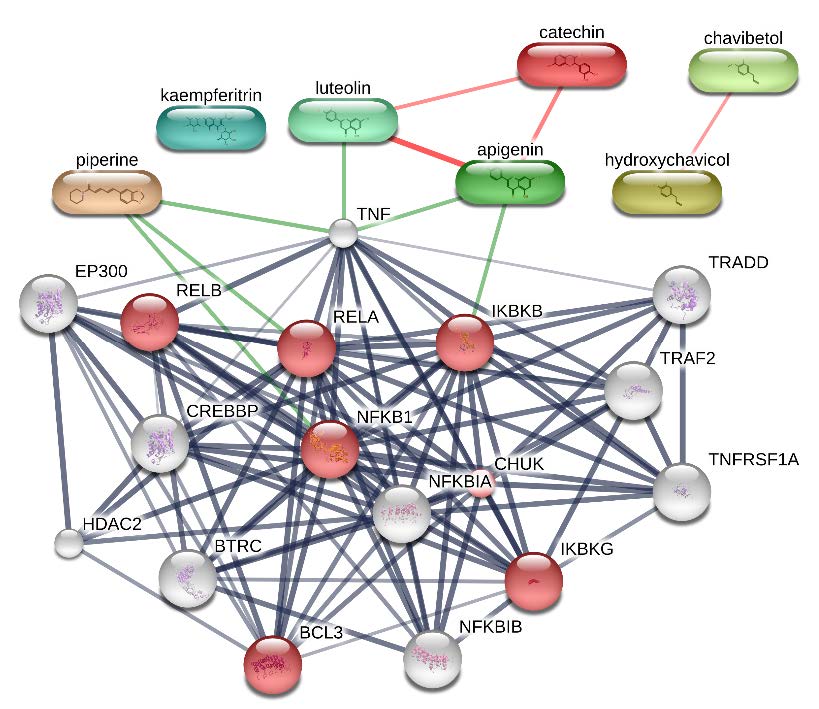
Figure 6. STITCH analysis between proteins involved in NFκB signaling pathway (red ball) and RBL active compounds.
DISCUSSION
RA is a systemic autoimmune disease that attacks the joints, thereby causing a decrease in the body's movement due to progressive functional impairment of the synovial joint. Meanwhile, TNF-α is a proinflammatory cytokine that plays an important role in the expression of the disease. It induces the activation and aggregation of the inflammatory cells, which causes the accumulation of immune cells at the site (Zhao et al., 2021). The binding on TNF-α to TNFR1 initiates the canonical pathway which can be activated by cytokines, growth factors, microbial components, stress agents, and mitogens thus activates NFκB signaling pathway. TNF-α and TNFR1 binding leads to an interaction with the IκB kinase (IKK) complex. IKK has two catalytic subunits, namely IKKα and IKKβ as well as a regulatory subunit called NFκB essential modulator (NEMO) (T. Liu et al., 2017). Furthermore, its activation induces the phosphorylation of IκB, which causes ubiquitination and degradation. Several studies reported that NFκB was involved in the pathogenesis of RA and it was found in the synovium of joints where it destroyed cartilages and bones.
RA is often treated with non-steroidal anti-inflammatory drugs (NSAIDs), which are used to reduce pain, fever, and other inflammatory processes. These drugs have antipyretic, anti-inflammatory as well as analgesic properties, and they include aspirin, ibuprofen, salsalate, mefenamic acid, and meloxicam. However, they have adverse effects on the gastric, renal, hepatic, and cardiovascular systems (Ghlichloo & Gerriets, 2022). Oral toxicity prediction with ProTox II also revealed that aspirin belongs to class III, which indicated that it is toxic. Alternative medications from the herbal plant have minimal adverse effects on the body, hence, RBL active compounds are safer than aspirin. Catechin was categorized in class VI, which indicates that it is non-toxic. It also belongs to the polyphenol group and several studies reported its bioactivities including anti-inflammation, anti-microbial, anti-virus, and anti-allergic (Bae et al., 2020).
In pharmacokinetics, distribution parameters consisted with VDss and BBB. VDss is considered low if the distribution volume value is below -0.15 log L/kg and high if it is above 0.45 log L/kg (Pires et al., 2015). VDss is a measurement of relative partitioning of drugs between serum and tissue based on the value of the partition coefficient (Singh & Singh, 2010). The partition coefficient is defined as the ratio of solute concentrations between two solvents namely lipophilic and hydrophilic. Lipophilic drugs are in the numerator and the hydrophilic drug is the denominator, then the logarithm value (log P) is calculated. Hydrophilic compounds with a low partition coefficient (below -0.15 logL/kg) were found mainly in areas that have high water content such as blood serum, whereas hydrophobic compounds with high partition coefficients (above 0.45 logL/kg) will be distributed to a hydrophobic region such as a lipid bilayer. RBL’s active compounds have a low distribution volume. It means that RBL’s active compounds were hydrophilic and found in the serum.
Drug excretion was determined based on total clearance and substrate OCT2. Kidney has a transporter to secrete drugs to the urine, namely organic cation transporters (OCTs). Organic cation transporters are important transponders for drug elimination from plasma and inside the kidneys. Drugs which act as substrate of OCT2 can cause OCT2 inhibition, thus the drug cannot be eliminated completely (Motohashi & Inui, 2013). Toxicity predictions used the Ames test. Ames test is a test to assess a compound that can be potentially mutagenic. Positive results indicate that the compounds are mutagenic and a carcinogen (Venkataramana et al., 2011).
Based on the molecular docking results, RBL active compounds have greater binding affinity than inhibitory drugs in TNFR1, NFκB p52/RelB/DNA, and IκK. The affinity value was determined with the Gibbs free energy (ΔG) in kcal/mol unit. ΔG value is negative when the docking system reaches equilibrium at a constant temperature and pressure (Balqis et al., 2022; Du et al., 2016). In the TNFR-ligands, the binding affinity of aspirin was -5.8 kcal/mol, while values of -9.8 kcal/mol and 8.2 kcal/mol were obtained from kaempferitrin and catechin, respectively. Aspirin (acetylsalicylic acid) is a non-steroidal medication, which is widely used as an anti-inflammatory drug. It also has several pharmacological activities including analgesic, antipyretic, and antiplatelet properties (Cadavid, 2017). Furthermore, several studies reported that it has numerous side effects in the gastrointestinal tract and also causes ulcer bleeding (Li et al., 2020).
The docking of NFκB p52/RelB/DNA-ligands revealed that the binding affinity value of kaempferitrin was -7.6 kcal/mol, which was also stronger than MG132 as an inhibitory drug (-6.8 kcal/mol). It is a proteasome inhibitor, which is involved in the modulation of inflammatory pain. Several studies explored the attenuating effect of MG132 on pain and joint inflammation in rat model RA (Ahmed et al., 2017). Kaempferitrin or kaempferol 3,7-dirhamnoside is a flavonoid glycoside that is mostly found in plants and is more abundant than flavonoid monomers. Flavonoid glycosides have various bioactivities including antidiabetic (Santos et al., 2019), antioxidant (Triches et al., 2004), and anti-inflammatory (Real-Sandoval et al., 2020).
A molecular docking study between IκK and ligands showed that apigenin (-8.3 kcal/mol) has the strongest binding affinity value among others including MG132. Apigenin is a secondary metabolite of plants and it belongs to the flavone class that is soluble in organic solvents. It has also been reported to have a beneficial function in alleviating the oxidative status and regulated pro-inflammatory expression in streptozotocin (STZ)-induced diabetic cardiomyopathy mice (H.-J. Liu et al., 2017). Furthermore, chavibetol, hydroxychavicol, and piperbetol are bioactive compounds present in glossy betel leaves, where they have various health benefits. They can modulate transcription factors and control reactive oxygen species (ROS) that are associated with cellular proliferation and pathways (Gundala & Aneja, 2014).
Based on amino acid residues on TNFR protein-ligands docking, serine-74 (Ser74) was present on all TNFR-ligand docking results (Table 7), which implies that it plays an important role for TNFR1. Saddala and Huang (2019) reported that there were several amino acid residues in TNFR1-TNF-α that play a significant role in the inhibitory mechanism of TNFR1. Those residues are Ser74, Ile58, Leu120, Gly121, Tyr515, Glu56, Ser57, Ser59, Cys73, Lys75, Arg77, Gln82, Cys96, Arg104, tyr106, and Asn110 (Saddala & Huang, 2019). The inhibitory mechanism on TNFR1 led to a decrease in the activation of NFκB. Meanwhile, molecular docking between NFκB p52/RelB/DNA complex with RBL active compounds and MG132 as drug inhibitors showed that serine 188 (Ser188) was present as a residue in all NFκB p52/RelB/DNA protein complex – ligands docking. Serine 222 was also present in most of the protein-ligand docking of NFκB p52/RelB/DNA protein complex (Table 7). This was likely caused by the activation of phosphorylate IκB, which inhibits NFκB at the two adjacent serine residues (Baichwal & Baeuerle, 1998; Mussbacher et al., 2019). Protein kinase was also phosphorylated at the residues of serine, threonine, and/or tyrosine (Pearlman et al., 2011). The inhibitory mechanism of RBL active compounds at serine residues causes a decrease in the phosphorylation that takes place at IκB. IκK is a protein kinase, which phosphorylates the IκB that are bonded to inactive NFκB. The molecular docking results in IκK showed that the residues of aspartic (Asp) and glutamic acid (Glu) play an important role in kinase activity. Furthermore, Cho et al. (2006) reported that glutamic acid was the most effective acidic amino acid for the phosphorylation mechanism of kinase activity, but aspartic acid also supports its mechanism of action (Cho et al., 2006).
Table7. Amino acid residue from molecular docking study.
|
Compounds |
TNFR1 |
NFκB p52/RelB/DNA complex |
IκK |
|||
|
Van der Waals |
Hydrogen Bond |
Van der Waals |
Hydrogen Bond |
Van der Waals |
Hydrogen Bond |
|
|
Aspirin |
The94, Val95, Asn110, Leu111, Phe112 |
Ser74, Asp93, Ser108, Glu109 |
- |
- |
- |
- |
|
MG132 |
- |
- |
Lys143, Met146, Lys182, Met185, Ser188, Asp257, Arg211, Gly314 |
Asp186, Leu187, Ser222, Gly224, Gln254, |
Gly22, Thr23, Gly24, Glu97, Tyr98, Glu100, Asp103, Lys147, Glu149, Asn150, Val152, Asp166, Leu167 |
Cys99 |
|
Apigenin |
Ser74, Lys75, Gln82, Asp93, Thr94, Val95, Arg104, Ser108, Glu109, Asn110, Phe112, |
Cys96 |
Met146, Lys183, Pro223 |
Tyr55, Ser188, Ser222 |
Leu21, Gly24, Ala42, Met96, Tyr98, Gly101, Gly102, Asp103 |
Glu100, Asp166 |
|
Catechin |
Ser72, Ser74, Lys75, Cys76, Arg77, Val95, Asn110, Phe112 |
Lys75, Arg77, Thr94, Cys96 |
His140, Lys183, Val184, Met185, Asp186, Ser222 |
Tyr55, Lys182, Ser188 |
Gly22, Gly24, Glu100, Gly102, Asp103, Lys147, Glu149, Asn150, Ile165 |
Thr23, Glu97, Cys99, Asp166 |
|
Chavibetol |
Ser74, Gln82, Thr94, Val95, Arg104, Glu109, Asn110, Phe112 |
Cys96 |
Ser222 |
Tyr55, His140, Ser188 |
Leu21, Thr23, Ala42, Glu97, Asp103, Glu149, Ele151, Val152 |
- |
|
Hydroxy-chavicol |
Ser74, Gln82, Val95, Arg104, Glu109, Asn110, Leu111 |
Thr94, Cys96 |
Asp186, Ser188 |
Met185 |
Ala42, Glu97, Asn150, Ile151, Val152 |
Asp103, Glu149 |
|
Kaempferitrin |
Leu71, Lys75, Cys76, Lys78, Met80, Gln82, Asp93, Asn11, Phe112 |
Ser72, Cys73, Ser74, Lys75,Cys76, Arg77, |
Tyr55, His140, Lys143, Lys183, Met185, Leu187, His218, Gly224, Gln254, Gly314 |
Ser188, Ser222, Arg311 |
Gly22, Ala42, Val74, Glu97, Cys99, Glu100, Gly101, Asp103, Lys147, Glu149, Asn150, Asp166, Gly184, Thr185. |
Thr23 |
|
Luteolin |
Ser74, Lys75, Met80, Asp93, Val95, Glu109, Asn110, Phe112 |
Gln82, Thr94, Ser108 |
Lys183, Asp186, Pro223 |
Tyr55, Lys182, Met185, Ser188, Ser222 |
Leu21, Thr23, Gly24, Lys44, Met96, Tyr98, Glu100, Gly101, Asp103, Asn150 |
Asp166 |
|
Piperbetol |
Cys73, Ser74, Lys75, Cys76, Arg77, Gln82, Asn110, Leu111 |
Ser72, Ser74, Arg77, Gln82 |
Tyr55, His140, Met146, Met185, Asp186, Ser188, Ser222, Pro223 |
- |
Lys44, Val74, Glu97, Tyr98, Gly102, Asp103, Glu149, Leu167, Gly168 |
Asp166 |
|
Piperine |
Ser74, Lys75, Cys76, Arg77, Gln82, Thr94, Arg104, Glu109, Asn110, Phe112 |
Cys96 |
His140, Lys143, Met146, Lys182, Lys183, Val184, Met185, Asp186 |
Tyr55, Ser188 |
Thr23, Gly24, Glu97, Tyr98, Cys99, Glu149, Asn150, Asp166 |
- |
The protein network interaction revealed the correlation between NFκB signaling and the TNF-mediated signaling pathway. The proteins involved in the pathways play a significant role in the canonical and non-canonical activation of NFκB. The canonical pathway relies on the activation of NKκB1, p50, RELA, and c-REL, while the non-canonical pathway activates the p100-sequestered NFκB members, namely p52 and RELB (Sun, 2017). Furthermore, the Canonical pathway depends on the degradation of IκB by IκK, while the non-canonical pathway depends on the phosphorylation-induced p100 processing and NFκB inducing kinase (NIK) (Sun, 2011).
This study results showed that RBL active compounds act as an inhibitor along the activation pathway. This starts with the inhibition of TNFR as a surface receptor, followed by IκK with its kinase activity, and then NFκB.
CONCLUSION
Based on the results, RBL active compounds have the potential of replacing synthetic drugs as an anti-inflammatory agent due to their efficacy and low toxicity. Catechin, kaempferitrin, and apigenin also have the potential due to their ability to inhibit the activation of NFκB. Additionally, further studies are advised on the in vitro and in vivo anti-inflammatory mechanism of these compounds.
AUTHOR CONTRIBUTIONS
Siti Imroatul Maslikah designed experiments, analysis and interpretation, supervised all the experiments, and writing the manuscript. Sri Rahayu Lestari, Nursasi Handayani, and Wira Eka Putra assisted in conducting the experiments and critical review. Alif Rofiqotun Nurul Alimah, Atikah Amalia, and Solichatul Afifah performed the data collections and data visualization. Siti Nur Arifah data collection and processing, writing the manuscript, and critical review. All authors have read and approved of the final manuscript.
CONFLICT OF INTEREST
The authors declare that they hold no competing interests.
REFERENCES
Ahmed, A.S., Ahmed, M., Li, J., Gu, H.F., Bakalkin, G., Stark, A., and Harris, H.E. 2017. Proteasome inhibitor MG132 modulates inflammatory pain by central mechanisms in adjuvant arthritis. International Journal of Rheumatic Diseases. 20: 25–32.
Alghasham, A. and Rasheed, Z. 2014. Therapeutic targets for rheumatoid arthritis: Progress and promises. Autoimmunity. 47: 77–94.
Almoallim, H., Al Saleh, J., Badsha, H., Ahmed, H.M., Habjoka, S., Menassa, J.A., and El-Garf, A. 2021. A review of the prevalence and unmet needs in the management of rheumatoid arthritis in Africa and the Middle East. Rheumatology and Therapy. 8: 1–16.
Alunno, A., Carubbi, F., Giacomelli, R., and Gerli, R. 2017. Cytokines in the pathogenesis of rheumatoid arthritis: New players and therapeutic targets. BMC Rheumatology. 1, 3.
Bae, J., Kim, N., Shin, Y., Kim, S.-Y., and Kim, Y.-J. 2020. Activity of catechins and their applications. Biomedical Dermatology. 4: 1-10.
Baichwal, V.R. and Baeuerle, P.A. 1998. Chapter 23 - Kinases in pro-inflammatory signal transduction pathways: new opportunities for drug discovery. In J. A. Bristol (Ed.), Annual Reports in Medicinal Chemistry (Vol. 33, pp. 233–242). Academic Press.
Balqis, B., Lukiati, B., Amin, M., Arifah, S.N., Atho’illah, M.F., and Widodo, N. 2022. Computational study of garlic compounds as potential anti-cancer agents for the inhibition of CCR5 and CXCR4. Chiang Mai University Journal of Natural Sciences. 21: e2022012.
Banerjee, P., Dehnbostel, F.O., and Preissner, R. 2018. Prediction is a balancing act: importance of sampling methods to balance sensitivity and specificity of predictive models based on imbalanced chemical data sets. Frontiers in Chemistry.
Banerjee, P., Eckert, A.O., Schrey, A.K., and Preissner, R. 2018. ProTox-II: A webserver for the prediction of toxicity of chemicals. Nucleic Acids Research. 46(Web Server issue), W257–W263.
Cadavid, A.P. 2017. Aspirin: The mechanism of action revisited in the context of pregnancy complications. Frontiers in Immunology. 8. https://www.frontiersin.org/article/10.3389/fimmu.2017.00261
Chang, M.J.W., Ko, C.Y., Lin, R.F., and Hsieh, L.L. 2002. Biological monitoring of environment exposure to safrole and the Taiwanese betel quid chewing. Archives of Environmental Contamination and Toxicology. 43: 432–437.
Chen, S., Feng, Z., Wang, Y., Ma, S., Hu, Z., Yang, P., Chai, Y., and Xie, X. 2017. Discovery of novel ligands for TNF-α and TNF Receptor-1 through Structure-based virtual screening and biological assay. Journal of Chemical Information and Modeling. 57: 1101–1111.
Cho, W.-H., Lee, Y.-J., Kong, S.-I., Hurwitz, J., and Lee, J.-K. 2006. CDC7 kinase phosphorylates serine residues adjacent to acidic amino acids in the minichromosome maintenance 2 protein. Proceedings of the National Academy of Sciences of the United States of America. 103: 11521–11526.
Dallakyan, S., and Olson, A.J. 2015. Small-molecule library screening by docking with PyRx. Methods in Molecular Biology (Clifton, N.J.). 1263: 243–250.
Dassault Systèmes BIOVIA. 2015. Discovery studio modeling environment, Version 4.5. Dassault Systèmes.
Drwal, M.N., Banerjee, P., Dunkel, M., Wettig, M.R., and Preissner, R. 2014. ProTox: A web server for the in silico prediction of rodent oral toxicity. Nucleic Acids Research. 42(Web Server issue), W53–W58.
Du, X., Li, Y., Xia, Y.-L., Ai, S.-M., Liang, J., Sang, P., Ji, X.-L., and Liu, S.-Q. 2016. Insights into protein–ligand interactions: Mechanisms, models, and methods. International Journal of Molecular Sciences. 17: 144.
Ghlichloo, I. and Gerriets, V. 2022. Nonsteroidal Anti-inflammatory Drugs (NSAIDs). In StatPearls. StatPearls Publishing. http://www.ncbi.nlm.nih.gov/books/NBK547742/
Gundala, S.R. and Aneja, R. 2014. Piper betel leaf: A reservoir of potential xenohormetic nutraceuticals with cancer-fighting properties. Cancer Prevention Research. 7: 477–486.
Gupta, S., Gupta, S.M., Sane, A.P., and Kumar, N. 2012. Chlorophyllase in Piper betle L. has a role in chlorophyll homeostasis and senescence dependent chlorophyll breakdown. Molecular Biology Reports. 39: 7133–7142.
Han, Y., Zhang, J., Hu, C.Q., Zhang, X., Ma, B., and Zhang, P. 2019. In silico ADME and toxicity prediction of ceftazidime and its impurities. Frontiers in Pharmacology. 10: 434.
Intriago, M., Maldonado, G., Cárdenas, J., and Ríos, C. 2019. Clinical characteristics in patients with rheumatoid arthritis: Differences between genders. The Scientific World Journal. 2019: e8103812.
Kadioglu, O., Nass, J., Saeed, M.E.M., Schuler, B., and Efferth, T. 2015. Kaempferol is an anti-inflammatory compound with activity towards NF-κB pathway proteins. Anticancer Research. 35: 6.
Kuhn, M., von Mering, C., Campillos, M., Jensen, L.J., and Bork, P. 2008. STITCH: Interaction networks of chemicals and proteins. Nucleic Acids Research, 36(Database issue), D684–D688.
Li, Z., Wang, Z., Shen, B., Chen, C., Ding, X., and Song, H. 2020. Effects of aspirin on the gastrointestinal tract: Pros vs. cons (Review). Oncology Letters. 20: 2567–2578.
Liu, H.-J., Fan, Y.-L., Liao, H.-H., Liu, Y., Chen, S., Ma, Z.-G., Zhang, N., Yang, Z., Deng, W., and Tang, Q.-Z. 2017. Apigenin alleviates STZ-induced diabetic cardiomyopathy. Molecular and Cellular Biochemistry. 428: 9–21.
Liu, T., Zhang, L., Joo, D., and Sun, S.-C. 2017. NF-κB signaling in inflammation. Signal Transduction and Targeted Therapy. 2: 1–9.
MacArthur, J., Bowler, E., Cerezo, M., Gil, L., Hall, P., Hastings, E., Junkins, H., McMahon, A., Milano, A., Morales, J., Pendlington, Z. M., Welter, D., Burdett, T., Hindorff, L., Flicek, P., Cunningham, F., and Parkinson, H. 2017. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Research, 45(Database issue), D896–D901.
Mirzaei, H., Zarbafian, S., Villar, E., Mottarella, S., Beglov, D., Vajda, S., Paschalidis, I. Ch., Vakili, P., and Kozakov, D. 2015. Energy minimization on manifolds for docking flexible molecules. Journal of Chemical Theory and Computation. 11: 1063–1076.
Motohashi, H. and Inui, K. 2013. Organic cation transporter OCTs (SLC22) and MATEs (SLC47) in the human kidney. The AAPS Journal. 15: 581–588.
Mussbacher, M., Salzmann, M., Brostjan, C., Hoesel, B., Schoergenhofer, C., Datler, H., Hohensinner, P., Basílio, J., Petzelbauer, P., Assinger, A., and Schmid, J. A. 2019. Cell type-specific roles of NF-κB linking inflammation and thrombosis. Frontiers in Immunology. 10: 85. https://www.frontiersin.org/article/10.3389/fimmu.2019.00085
Okada, Y., Wu, D., Trynka, G., Raj, T., Terao, C., Ikari, K., Kochi, Y., Ohmura, K., Suzuki, A., Yoshida, S., Graham, R.R., Manoharan, A., Ortmann, W., Bhangale, T., Denny, J.C., Carroll, R.J., Eyler, A.E., Greenberg, J.D., Kremer, J.M., … Plenge, R.M. 2014. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 506: 376–381.
Pearlman, S.M., Serber, Z., and Ferrell, J.E. 2011. A mechanism for the evolution of phosphorylation sites. Cell. 147: 934–946.
Pires, D.E.V., Blundell, T.L., and Ascher, D.B. 2015. pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. Journal of Medicinal Chemistry. 58: 4066–4072.
Real-Sandoval, S.A., Gutiérrez-López, G.F., Domínguez-López, A., Paniagua-Castro, N., Michicotl-Meneses, M.M., and Jaramillo-Flores, M.E. 2020. Downregulation of proinflammatory liver gene expression by Justicia spicigera and kaempferitrin in a murine model of obesity-induced by a high-fat diet. Journal of Functional Foods. 65: 103781.
Saddala, M.S. and Huang, H. 2019. Identification of novel inhibitors for TNFα, TNFR1 and TNFα-TNFR1 complex using pharmacophore-based approaches. Journal of Translational Medicine. 17: 215.
Safiri, S., Kolahi, A.A., Hoy, D., Smith, E., Bettampadi, D., Mansournia, M.A., Almasi-Hashiani, A., Ashrafi-Asgarabad, A., Moradi-Lakeh, M., Qorbani, M., Collins, G., Woolf, A.D., March, L., and Cross, M. 2019. Global, regional and national burden of rheumatoid arthritis 1990-2017: A systematic analysis of the Global Burden of Disease study 2017. Annals of the Rheumatic Diseases. 78: 1463–1471.
Santos, M., Fortunato, R.H., and Spotorno, V.G. 2019. Analysis of flavonoid glycosides with potential medicinal properties on Bauhinia uruguayensis and Bauhinia forficata subspecies pruinosa. Natural Product Research. 33: 2574–2578.
Sarma, C., Rasane, P., Kaur, S., Singh, J., Singh, J., Gat, Y., Garba, U., Kaur, D., and Dhawan, K. 2018. Antioxidant and antimicrobial potential of selected varieties of Piper betle L. (Betel leaf). Anais Da Academia Brasileira de Ciências. 90: 3871–3878.
Singh, Y.P., and Singh, R.A. 2010. In silico studies of organosulfur-functional active compounds in garlic. BioFactors. 36: 297–311.
Smolen, J.S., Aletaha, D., and McInnes, I. B. 2016. Rheumatoid arthritis. Lancet (London, England), 388(10055): 2023–2038.
Sun, S.-C. 2011. Non-canonical NF-κB signaling pathway. Cell Research. 21: 71–85.
Sun, S.-C. 2017. The non-canonical NF-κB pathway in immunity and inflammation. Nature Reviews Immunology. 17: 545–558.
Szklarczyk, D., Franceschini, A., Wyder, S., Forslund, K., Heller, D., Huerta-Cepas, J., Simonovic, M., Roth, A., Santos, A., Tsafou, K. P., Kuhn, M., Bork, P., Jensen, L.J., and von Mering, C. 2015. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Research, 43(Database issue). D447-452.
Triches, E., Zanatta, L., Seifriz, I., Creczynski-Pasa, tânia, Pizzolatti, M., Szpoganicz, B., and Silva, F. 2004. Hypoglycemic effect and antioxidant potential of kaempferol-3,7- o -(??)-dirhamnoside from Bauhinia forficata leaves. Journal of Natural Products. 67: 829–832.
Trott, O., and Olson, A. J. 2010. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. Journal of Computational Chemistry. 31: 455–461.
Tsubaki, M., Takeda, T., Kino, T., Itoh, T., Imano, M., Tanabe, G., Muraoka, O., Satou, T., and Nishida, S. 2015. Mangiferin suppresses CIA by suppressing the expression of TNF-α, IL-6, IL-1β, and RANKL through inhibiting the activation of NF-κB and ERK1/2. American Journal of Translational Research. 7: 1371–1381.
Venkataramana, C.H.S., Sravani, K.M.R., Singh, S.S., and Madhavan, V. 2011. In-silico ADME and toxcity studies of some novel indole derivatives. Journal of Applied Pharmaceutical Science, 01(10): 159–162.
Zhao, X., Kim, Y.-R., Min, Y., Zhao, Y., Do, K., and Son, Y.-O. 2021. Natural plant extracts and compounds for rheumatoid arthritis therapy. Medicina. 57: 266.
OPEN access freely available online
Natural and Life Sciences Communications
Chiang Mai University, Thailand.
Siti Imroatul Maslikah1,*, Sri Rahayu Lestari1, Nursasi Handayani1, Wira Eka Putra1,2, Alif Rofiqotun Nurul Alimah3, Atikah Amalia1, Solichatul Afifah1, and Siti Nur Arifah1
1 Department of Biology, Faculty of Mathematics and Natural Sciences, Universitas Negeri Malang, Indonesia.
2 Department of Biotechnology, Faculty of Mathematics and Natural Sciences, Universitas Negeri Malang, Indonesia.
3 Department of Biology, Faculty of Mathematics and Natural Sciences, Universitas Brawijaya, Indonesia.
Corresponding author: Siti Imroatul Maslikah, E-mail: siti.imroatul.fmipa@um.ac.id
Total Article Views
Editor: Pachara Sattayawat,
Chiang Mai University, Thailand
Article history:
Received: August 26, 2022;
Revised: October 13, 2022;
Accepted: October 21, 2022;
Published online: November 1, 2022

