
Synthesis, Molecular Docking, and ADMET Study of N1-Hydrogen and N1-Benzoyl Pyrazoline as Antibacterial Agents
Putra Jiwamurwa Pama Tjitda, Jumina and Tutik Dwi Wahyuningsih*Published Date : 2022-07-11
DOI : https://doi.org/10.12982/CMUJNS.2022.048
Journal Issues : Number 3, July-September 2022
Abstract Syntheses of N1-hydrogen and N1-benzoyl pyrazoline derivatives and their antibacterial in vitro and in silico assays have been carried out. N1-Hydrogen pyrazoline derivatives were synthesized by cyclization of 2’-hydroxy chalcone, and the subsequent substitution reaction produced N1-benzoyl pyrazoline derivatives. The in vitro antibacterial assay was carried out by disc diffusion method. In silico evaluation was performed via molecular docking against ecKAS III enzyme (ID PDB: 1hnj) and ADMET prediction was carried out using pkCSM tool. The synthesis results showed that N1-hydrogen and N1-benzoyl pyrazoline derivatives were yielded in 50-83%. Antibacterial test results indicated that the presence of N1-benzoyl substituent decreased the antibacterial activity and was only active on Gram-positive bacteria. In comparison, the N1-hydrogen pyrazolines exhibited good antibacterial activity against both Gram-positive and negative bacteria. The ADMET result confirms that compound 2 has the potential to be evolved as a drug in the future.
Keywords: Pyrazoline, Antibacterial, Molecular Docking, ADMET
Citation: Tjitda, P.J.P., Jumina, and Wahyuningsih, T.D. 2022. Synthesis, molecular docking, and ADMET study of N1-Hydrogen and N1-Benzoyl Pyrazoline as antibacterial agents. CMUJ. Nat. Sci. 21(3): e2022048.
INTRODUCTION
The infection caused by some bacteria, fungi, and parasites has been a major threat to human health in the past few years. Although many commercial drugs are available in the marketplace, drug resistance cases toward bacteria, fungi, and parasites have been reported (Li and Webster, 2018). The one solution that could be offered is to design and develop new active compounds in various chemical synthetic approaches to solve the drug resistance as well as work up the activity of the drug.
One of the bioactive synthetic compounds is pyrazoline. In reported literature, pyrazoline derivatives have broad bioactivities, i.e., antimicrobial (Desai et al., 2017), antioxidant (Rana et al., 2021), anti-inflammatory (Fernandes et al., 2017), aanalgesic (Abeed et al., 2019), antitumor (Alkamaly et al., 2021), and antibacterial (Pola et al., 2020).
The synthesis of pyrazoline compound involves Fischer and Knoevenagel reaction as cyclizations reaction between hydrazine (Hoz et al., 2021) or phenylhydrazine (Jadhav et al., 2018) and chalcone. The 2'-hydroxy chalcone is one of the chalcone derivatives with outstanding activity as an antibacterial agent (Freitas et al., 2020). On the other hand, vanillin, an abundant natural product in Indonesia, is generally discovered from the extraction of vanilla pods and used to flavoring in food. Illicachi et al. (2017) have successfully utilized vanillin to synthesize chalcone derivatives through an aldol condensation reaction between vanillin and acetophenone derivatives.
Some research concerning the modification of the pyrazoline ring has been reported. The administration of some substituents on the N1-position shows the improvement of biological activity. The presence of a carboxamide group (-CONH2) on the N1 pyrazoline ring contributes to antimicrobial activity (Azarifar and Shaebanzadeh, 2002). De and Saha (2019) have successfully modified the pyrazoline ring using phenyl on the N1 position. The synthetic product showed good antimicrobial in vitro. Baseer et al. (2013) reported that acetylated pyrazoline has better antibacterial activity than non-acetylated pyrazoline.
In addition, Patel et al. (2013) reported that benzoyl substituted on N1 pyrazoline ring also increases antibacterial activity with inhibition zone to be 10-27 mm. They declared that the pyrazoline derivatives contribute to the pharmacological character, particularly as an antibacterial agent. N1 position of pyrazoline ring is attractively one thing for developing new compounds with excellent antibacterial.
The β-ketoacyl-acyl carrier protein (ACP) synthase III (ecKAS III) is an enzyme that is liable to the persistence of fatty acid biosynthesis, which is the main component in membrane cells (Qiu et al., 2001). When ecKAS III activity is blocked, it is the potential to interfere stability of membrane cells, and the sustainability of bacteria cells is disturbed. The emergence of benzoyl on the N1 position of the pyrazoline ring's N1 position will hopefully enhance the lipophilicity character that affects its antibacterial activity.
Under the explanation above, this research aims to synthesize N1-benzoyl pyrazoline derivatives by taking vanillin as feedstock. As a comparison, this study discusses the synthesis of N1-hydrogen pyrazoline, which constitutes a precursor for producing N1-benzoyl pyrazoline. All synthetic product is examined to evaluate their antibacterial activity. Molecular docking is carried out on all synthetic compounds to evaluate model interaction and binding energy. The absorption, distribution, metabolism, excretion, and toxicity (ADMET) of all compounds are also studied through an in silico study. This study is expected to provide information for N1-substituted Pyrazoline-based drug development further.
MATERIAL AND METHODS
Materials
All of the materials were purchased from Merck with pro analysis grade. The melting point was determined using melting point apparatus (Electrothermal-9100), the functional group measured by Spectrometer Fourier transform infrared (FTIR, Shimadzu Prestige 21), the purity and mass of compound recorded on Gas Chromatograph-Mass Spectrometer (GC-MS, Shimadzu QP 2010S), the proton and carbon spectra obtained from Nuclear Magnetic Resonance (NMR 1H (500 MHz) and 13C (125 MHz)), respectively. Molecular docking of the synthetic compound was carried out using AutoDock Vina. The 2D and 3D structures were generated using ChemDraw Professional 16, and the structure optimization was done using Gaussian 09 software, while the molecule interaction with protein was visualized using Discovery Studio software.
Synthesis of 1-(2-hydroxyphenyl)-3-(4-hydroxy-3-methoxyphenyl)-2-propen-1-on
(Chalcone 1)
Synthesis of chalcone 1 was performed by following the previous method (Kumar et al., 2013) with slight modification. 2’-Hydroxy acetophenone (5 mmol) was added into 2.5 mL sodium hydroxide (40%, b/v) in methanol. Afterward, vanillin (5 mmol) was added dropwise into the mixture. The reaction proceeded for 4 h and was monitored using a thin layer chromatography (TLC). The mixture was poured into the crushed ice and then neutralized using an acid solution to get the yellow precipitate. The precipitate was filtered off, washed with cooled water, and dried. The purified product was obtained by recrystallization, and this product was characterized by FTIR, GC-MS, 1H- and 13C-NMR. This method was also used for synthesizing chalcone 2 by replacing vanillin with veratraldehyde.
1-(2-hydroxyphenyl)-3-(4-hydroxy-3-methoxyphenyl)-2-propen-1-on
(Chalcone 1)
Yield: 55.39% (light yellow solid), melting point: 116-118°C, FTIR (KBr, cm-1): 3403, 3055, 1635, 1512, 1581, 1442, 1026, 987; 1H-NMR (CDCl3, δ): 3.98 (s, 3H, OCH3), 5.94 (s, 1H, OH ArB), 6.94 (m, 1H, ArB), 6.98 (d, 1H, ArB, J=8.45 Hz), 7.03 (dd, 1H, ArB, J=1.30 and 1.30 Hz), 7.15 (d, 1H, ArA, J=1.95 Hz), 7.27 (d, 1H, ArA, J=1.95 Hz), 7.49 (dd, 1H, ArA, J=1.30 and 1.95 Hz), 7.51 (d, 1H, Hα, J=15.00 Hz), 7.88 (d, 1H, Hβ, J=15.00 Hz), 7.93 (dd, 1H, ArA, J=1.30 and 1.95 Hz), 12.91 (1H, OH, ArA); 13C-NMR (CDCl3, δ): 193.77 (C=O), 163.73 (C-2’), 148.85 (C-3), 147.01 (Cβ), 146.02 (C-4), 136.37 (C-4’), 129.70 (C-6’), 127.41 (C-1), 123.94 (C-1’), 120.28 (C-5’), 118.93 (Cα), 118.81 (C-6), 117.69 (C-5), 115.14 (C-3’), 110.41 (C-2), 56.25 (C-OCH3). Massa spectrum (m/z): 270 (M+), 239, 150, 121, 93.
1-(2-hydrixyphenyl)-3-(3,4-dimethoxyphenyl)-2-propen-1-on
(Chalcone 2)
Yield: 77.97% (light yellow solid), melting point: 87-90°C, FTIR (KBr, cm-1): 3448, 3078, 1635, 1512, 1560, 1442, 1026, 995; 1H-NMR (CDCl3, δ): 3.90 (s, 3H, OCH3), 3.92 (s, 3H, OCH3), 3.94 (s, 3H, OCH3), 3.96 (s, 3H, OCH3), 6.92 (m, 3H, ArB), 7 (d, 1H, ArA, J=1.95 Hz), 7.27 (dd, 1H, ArA, J=1.95 and 1.95 Hz), 7.49 (m, 2H, ArA), 7.52 (d, 1H, Hα, J=15.00 Hz), 7.88 (d, 1H, Hβ, J=15.00 Hz), 7.93 (dd, 2H, ArA, J=1.95 and 1.30 Hz), 12.90 (s, 1H, OH ArA); 13C-NMR (CDCl3, δ): 192.33 (C=O), 161.70 (C-2’), 149.57 (C-3), 149.40 (C-4), 136.36 (Cβ), 129.70 (C-4’), 127.20 (C-6’), 123.80 (C-1), 121.77 (C-1’), 120.24 (C-5’), 118.91 (Cα), 118.77 (C-6), 117.91 (C-3’), 111.26 (C-5), 109.52 (C-2), 56.11 (C-OCH3). Massa spectrum (m/z): 284 (M+), 253, 164, 121, 91.
Synthesis of 1-hydrogen-3-(2-hydroxyphenyl)-5-(4-hydroxy-3-methoxyphenyl)-2-pyrazoline
(N1-hydrogen pyrazoline A)
Synthesis of N1-hydrogen pyrazoline was carried out following the method by Setyawati et al. (2017) with slight modification. The mixture of chalcone 1 (1 mmol) and hydrazine monohydrate (3 mmol) in methanol was dissolved and refluxed for 2 h. The mixture was poured into the crushed ice to give a precipitate; then, it was filtered, washed, and dried. The purity of the product was analyzed by TLC using n-hexane: ethyl acetate (7:3) as eluent. The product of N1-hydrogen pyrazoline was purified by recrystallization, and then it was characterized using FTIR, GC-MS, 1H- and 13C-NMR. This procedure was applied to the synthesis of compound 2 by replacing chalcone 1 with chalcone 2.
1-hydrogen-3-(2-hydroxyphenyl)-5-(4-hydroxy-3-methoxyphenyl)-2-pyrazoline
(Compound 1)
Yield: 76.22% (dark yellow solid), melting point: 128-130°C, FTIR (KBr, cm-1): 3379, 3302, 3066, 1597, 1041; 1H-NMR (CDCl3, δ): 3.10 (dd, pyrazoline Hα, J=9.75 and 9.70 Hz), 3.54 (dd, pyrazoline Hβ, J= 11.00 and 10.40 Hz), 3.88 (s, 3H, OCH3), 4.83 (t, pyrazoline Hx), 5.62 (s, 1H, OH ArB), 5.96 (s, 1H, NH), 6.85 (m, 1H, ArB), 6.88 (d, 2H, ArB, J=8.45 Hz), 6.92 (d, 1H, ArA, J=1.95 Hz), 7.01 (d, 1H, ArA, J=7.80 Hz), 7.16 (dd, 1H, ArA, J=1.30 and 1.30 Hz), 7.25 (dd, 1H, ArA, J=1.30 and 1.30 Hz), 11.02 (s, 1H, OH ArA); 13C-NMR (CDCl3,δ): 157.91 (C-2’), 154.65 (C=N), 147.01 (C-3), 145.62 (C-4), 133.90 (C-1), 130.62 (C-4’), 127.75 (C-6’), 119.71 (C-5’), 119.28 (C-6), 116.73 (C-1’), 116.63 (C-5), 114.67 (C-3’), 108.68 (C-2), 62.98 (C-OCH3), 56.10 (CH-NH), 41.98 (CH2). Mass spectrum (m/z): 284 (M+), 253, 160, 135, 107, 77, 51.
1-hydrogen-3-(2-hydroxyphenyl)-5-(3,4-dimethoxyphenyl)-2-pyrazoline
(Compound 2)
Yield: 84.39% (brownish gray solid), melting point: 105-108°C, FTIR (KBr, cm-1): 3394, 3348, 3066, 1589, 1026; 1H-NMR (CDCl3, δ): 3.11 (dd, pyrazoline Hα, J=9.70 and 9.05 Hz), 3.55 (dd, pyrazoline Hβ, J= 11.00 and 10.40 Hz), 3.87 (s, 3H, OCH3), 4.85 (t, pyrazoline Hx), 5.98 (s, 1H, NH), 6.83 (d, 1H, ArB, J=7,80 Hz), 6.89 (m, 2H, ArB,), 6.93 (d, 1H, ArA, J=1.95 Hz), 7.01 (d, 1H, ArA, J=7.15 Hz), 7.17 (dd, 1H, ArA, J=1.30 and 1.30 Hz), 7.25 (dd, 1H, ArA, J=1.30 and 1.30 Hz), 11.04 (s, 1H, OH ArA); 13C-NMR (CDCl3, δ): 157.92 (C-2’), 154.60 (C=N), 149.50 (C-3), 148.97 (C-4), 134.47 (C-1), 130.62 (C-4’), 127.74 (C-6’), 119.27 (C-5’), 118.88 (C-6), 116.77 (C-1’), 116.62 (C-5), 111.41 (C-3’), 109.42 (C-2), 62.90 (2xC-OCH3), 56.11 (CH-NH), 41.92 (CH2). Mass spectrum (m/z): 298 (M+), 267, 161, 135, 107, 77, 51.
Synthesis of 1-benzoyl-3-(2-hydroxyphenyl)-5-(4-hydroxy-3-methoxyphenyl)-2-pyrazoline
(Compound 3)
Synthesis of N1-hydrogen pyrazoline refers to the method obtained by Suma et al. (2017) with slight modification. Compound 1 (0.5 mmol) in methanol was added to 5 mL glacial acetic acid. Subsequently, 1 mmol of benzoyl chloride was poured into the mixture. The mixture was then refluxed over 2 h, and it was monitored by TLC. After completing the reaction, the mixture was poured into crushed ice. The precipitate was further filtered off, washed with fresh water, and dried in the desiccator. The final product was purified by recrystallization using methanol. The structure characterization was conducted using FTIR, GC-MS, 1H- and 13C-NMR. This method was also used for synthesizing compound 4 by replacing compound 1 with compound 2.
1-benzoyl-3-(2-hydroxyphenyl)-5-(4-hydroxy-3-methoxyphenyl)-2-pyrazoline
(Compound 3)
Yield: 55.72% (brownish white solid), melting point: 198-200°C, FTIR (KBr, cm-1): 3448, 3232, 1620, 1597, 1033; 1H-NMR (CDCl3, δ): 3.36 (dd, pyrazoline Hα, J=5.20 and 5.20 Hz), 3.88 (s, 3H, OCH3), 3.89 (dd, pyrazoline Hβ, J=11.05 and 5.20 Hz), 5.64 (s, 1H, OH ArB), 5.70 (dd, pyrazoline Hx, J=5.20 and 5.15 Hz), 6.89 (m, 3H, ArB, 1H, ArA), 6.99 (d, 1H, ArA, J=8.40 Hz), 7.23 (dd, 1H, ArA, J=1.95 and 1.95 Hz), 7.33 (m, 1H, ArA), 7.49 (m, 3H, ArC), 7.79 (d, 2H, ArC, J=7.10 Hz), 10.0 (s, 1H, OH ArA); 13C-NMR (CDCl3, δ): 167.08 (C=O), 157.71 (C-2’), 156.75 (C=N), 146.88 (C-3), 145.57 (C-4), 134.20 (C-1), 133.27 (C-1’’), 132.53 (C-4’), 131.43 (C-4’’), 128.86 (C-6’), 128.60 (C-3’’, C-5’’), 128.31 (C-2’’, C-6’’), 121.50 (C-5’), 119.90 (C-6), 118.55 (C-1’), 117.27 (C-5), 115.15 (C-3’), 108.99 (C-2), 56.10 (C-OCH3), 42.49 (CH-N), 29.89 (CH2).
1-benzoyl-3-(2-hydroxyphenyl)-5-(3, 4-dimethoxyphenyl)-2-pyrazoline
(Compound 4)
Yield: 82.49% (light brown solid), melting point: 110-113°C, FTIR (KBr, cm-1): 3325, 3055, 1681, 1589, 1026; 1H-NMR (CDCl3, δ): 3.36 (dd, pyrazoline Hα, J=5.85 and 5.20 Hz), 3.85 (s, 3H, OCH3), 3.88 (s, 3H, OCH3), 3.89 (dd, pyrazoline Hβ, J=11.70 and 11.65 Hz), 5.70 (dd, pyrazoline Hx, J=5.20 and 5.15 Hz), 6.85 (d, 1H, ArB, J=8.40 Hz), 6.91 (m, 2H, ArB, 1H, ArA), 6.99 (d, 1H, ArA, J=7.10 H), 7.23 (dd, 1H, ArA, J=1.95 and 1.95 Hz), 7.33 (m, 3H, ArA), 7.49 (m, 3H, ArC), 7.49 (d, 2H, ArC, J=7.10 Hz), 10,09 (s, 1H, OH ArA); 13C-NMR (CDCl3, δ): 167.08 (C=O), 157.72 (C-2’), 156.71 (C=N), 149.53 (C-3), 148.96 (C-4), 136.80 (C-1), 134.20 (C-1’’), 132.53 (C-4’), 131.43 (C-4’’), 128.87 (C-6’), 128.59 (C-3’’, C-5’’), 128.32 (C-2’’, C-6’’), 119.90 (C-5’), 117.97 (C-6), 117.27 (C-1’), 116.00 (C-5), 111.79 (C-3’), 109.43 (C-2), 59.31 (C-OCH3), 56.14 (C-OCH3), 42.46 (CH-N), 29,88 (CH2). Mass spectrum (m/z): 401 (M+), 297, 165, 120, 105, 77, 51.
Antibacterial Activity
All synthesized compounds are assessed for antibacterial activity against Gram-positive bacteria such as Staphylococcus aureus, Bacillus cereus, and Bacillus subtilis, while Gram-negative bacteria are Escherichia coli and Shigella flexneri. The antibacterial assay uses the Kirby-Bauer disc diffusion method. The antibacterial activity of the synthetic compound was evaluated by the diameter of the inhibition zone.
Molecular Docking
The 2D structures of synthetic compounds were sketched and converted to 3D formats. The optimization of the 3D structure with Austin Model 1 (AM1) method using Gaussian 09 software was then performed. All synthetic compounds were docked to ecKAS III enzyme, which was retrieved from the RSCB website with ID PDB: 1hnj (1.46 Å). The docking method was validated and determined using the Root Mean Square Deviation (RMSD) value under 2 (Da Silva Costa et al., 2018). Molecular docking of the entire compound was executed using AutoDock Vina (Trott and Olson, 2009). The binding energy value was shown in a notepad, and visualization of binding and type interaction was performed using Discovery studio software.
ADMET Study
The whole synthetic compounds were predicted adsorption, distribution, metabolism, excretion, and toxicity (ADMET) by in silico study. These characters describe the pharmacokinetics of drugs, and its result could be a reference for producing drug candidates in the future. This prediction used the pkCSM tool (Pires et al., 2015).
RESULTS
Synthesis of N1-benzoyl pyrazoline compound
Syntheses of N1-benzoyl compounds involve several reactions. several reactions. Firstly, the synthesis of chalcone compound from aromatic aldehyde derivatives and 2’-hydroxy acetophenone. Chalcone, a precursor compound, is used as an electrophilic source to produce N1-hydrogen pyrazoline. This reaction reacts to benzoyl chloride by The SN2 reaction to yield N1-benzoyl pyrazoline. Overall, the reaction N1-benzoyl pyrazoline synthesis is provided in Figure 1.
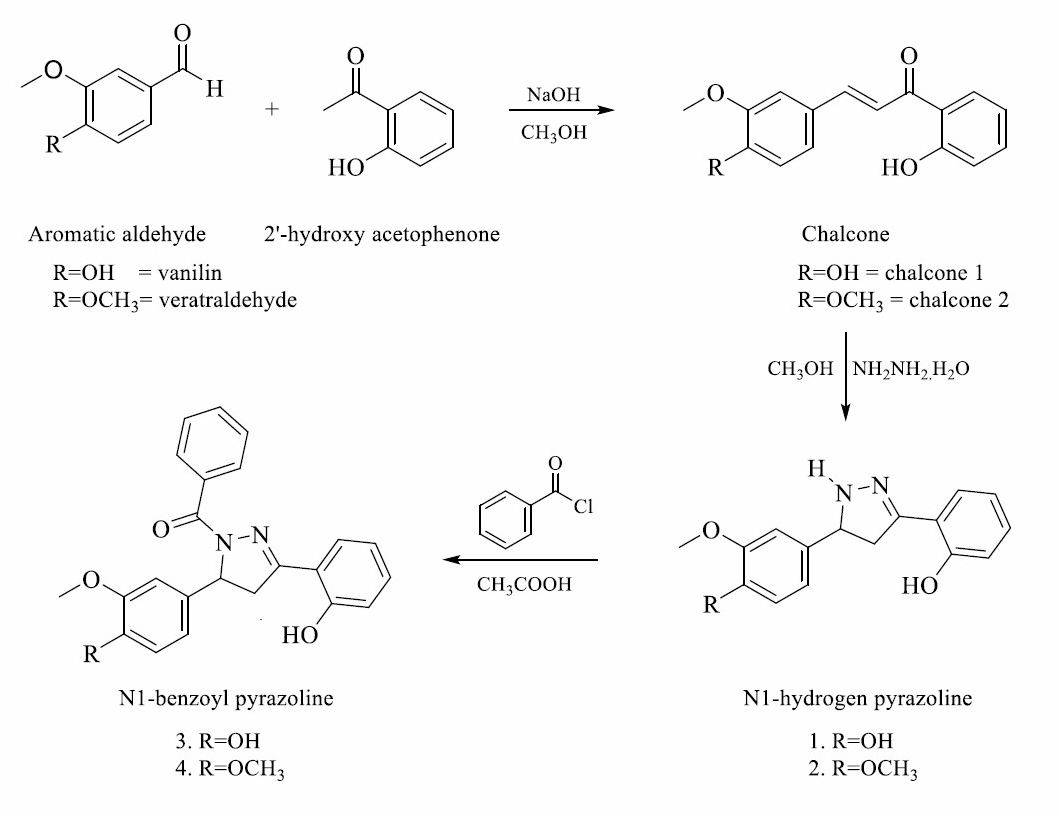
Figure 1. The scheme of synthesis reaction of N1-hydrogen and N1-benzoyl pyrazoline derivatives.
Antibacterial activity
As shown in Table 1, the antibacterial activity was performed on all compounds and the examination was conducted on five bacteria. The three bacteria are coming from Gram-positive and two bacteria from Gram-negative.
Table 1. Antibacterial activity of synthesized compounds.
|
Compound |
Diameter of inhibition zone (mm) |
||||
|
Staphylococcus aureus |
Bacillus cereus |
Bacillus subtilis |
Escherichia coli |
Shigella flexneri |
|
|
1 |
9.35 |
11.00 |
4.30 |
7.75 |
8.75 |
|
2 |
7.20 |
3.00 |
3.00 |
5.25 |
3.75 |
|
3 |
2.45 |
2.75 |
2.00 |
- |
- |
|
4 |
- |
3.25 |
1.75 |
- |
- |
|
DMSO (negative control) |
- |
- |
- |
- |
- |
|
Tetracycline (positive control) |
16.00 |
19.25 |
7.25 |
16.25 |
17.00 |
Molecular docking
Molecular docking of all synthetic compounds to the ecKAS III enzyme was performed. In this work, the validation of the docking protocol was carried out to assure the docking procedure could be adapted to the entire testing ligand. The RMSD re-docking value produced below 2 Å. Figure 2 depicts the superimposed structure of the MLC (Malonyl-coenzyme A) as a native ligand (green) into native ligand re-docking (blue). The resulting RMSD value gives 0.963 Å.
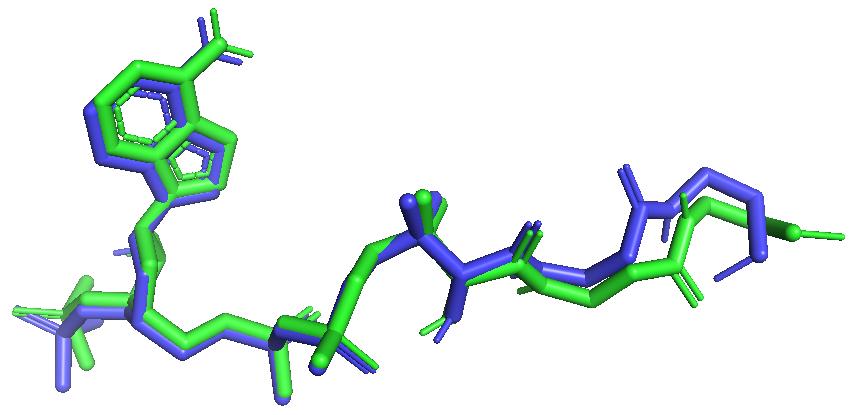
Figure 2. RMSD analysis result of native ligand re-docking to ecKAS III enzyme (ID PDB: 1hnj).
Molecular docking of the whole testing ligand toward enzyme ecKAS III presented the results of binding energy and type of interaction. The resulting docking data are summarized in Table 2.
Table 2. Binding energy and interaction of docking result.
|
Compound |
Binding energy (kcal/mol) |
Amino acid residue |
Interaction |
|
|
Type |
Distance (Amstrong) |
|||
|
MLC |
-8.4 |
Asn247 |
Hydrogen Bond |
3.09 |
|
|
|
Gly209 |
Hydrogen Bond |
2.63 |
|
|
|
Thr28 |
Hydrogen Bond |
2.42 |
|
|
|
His244 |
Pi-sulfur |
- |
|
|
|
Arg36 |
Electrostatic |
- |
|
1 |
-8.2 |
Asn274 |
Hydrogen bond |
3.13 |
|
|
|
Asn247 |
Hydrogen bond |
4.17 |
|
|
|
Cys112 |
Hydrophobic |
- |
|
2 |
-8.1 |
Asn247 |
Hydrogen bond |
2.67 |
|
|
|
Asn274 |
Hydrogen bond |
3.22 |
|
|
|
Ala246 |
Hydrogen bond |
2.51 |
|
|
|
Arg36 |
Hydrogen bond |
2.58 |
|
|
|
Cys112 |
Hydrophobic |
- |
|
3 |
-8.3 |
Asn247 |
Hydrogen bond |
3.07 |
|
4 |
-8.3 |
ASn247 |
Hydrogen bond |
3.10 |
To identify the interaction of testing ligands on the active side of ecKAS III enzyme, visualization of both 2- and 3-dimension interaction of all testing ligands was executed. Also, the interaction of native ligand in the pocket of ecKAS III enzyme was displayed in Figure 3.
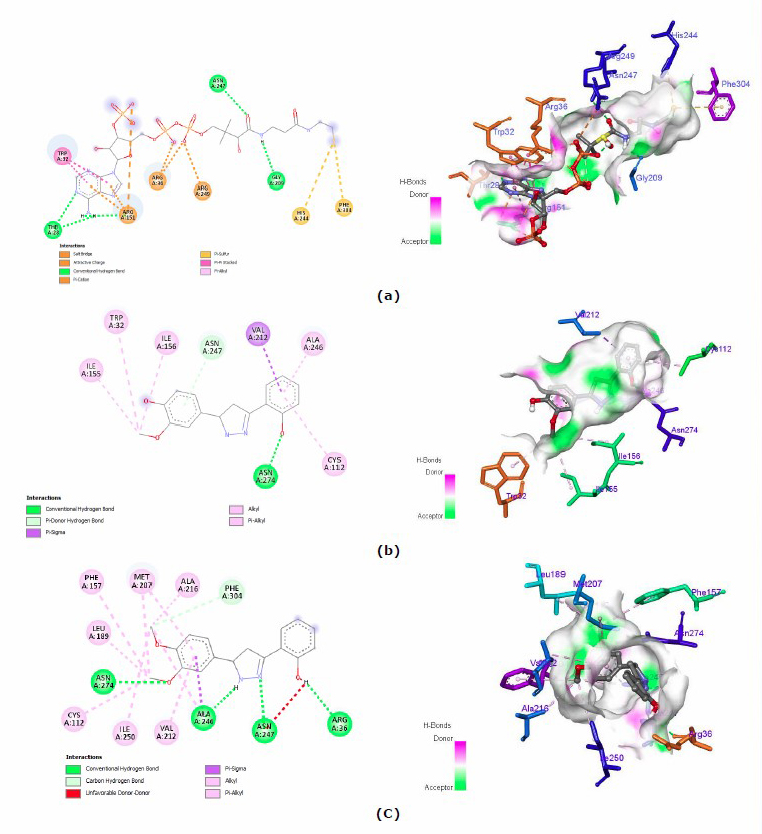
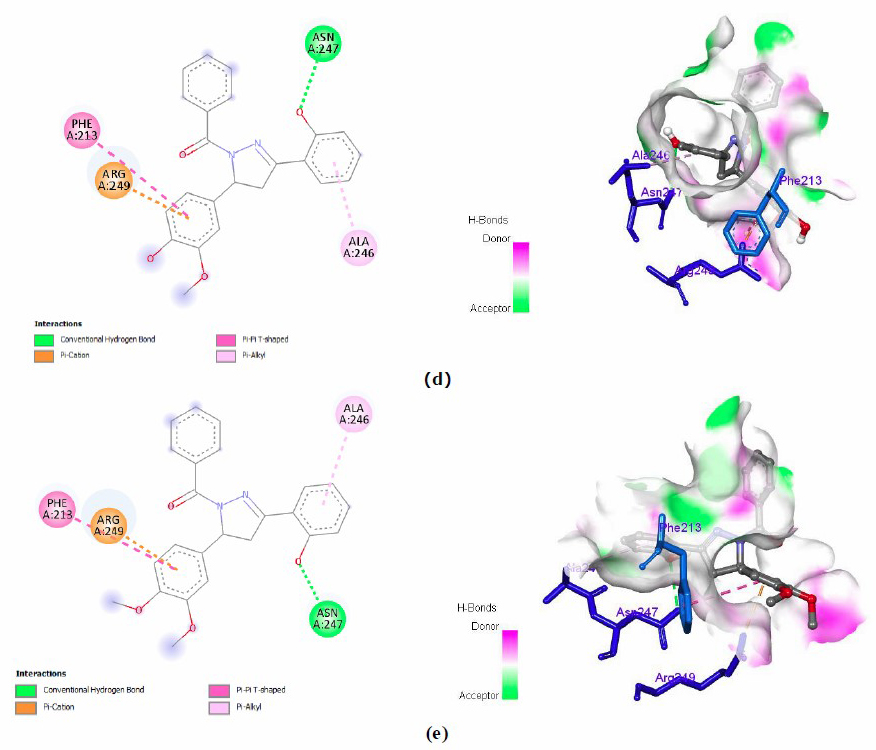
Figure 3. 2D and 3D interactions of a) native ligand, b) compound 1, c) compound 3, d) compound 3, and e) compound 4.
ADMET study
The pharmacokinetic character of synthetic compounds had been studied by in silico analysis through ADMET property. Each simplified molecular-input line-entry system (SMILE) code of synthetic compound was entered on the pkCSM website, and the obtained result was displayed in Table 3.
Table 3. ADMET characteristic of all compounds.
|
|
Compound 1 |
Compound 2 |
Compound 3 |
Compound 4 |
|
Absorption |
|
|
|
|
|
Intestinal absorption (%) |
91.119 |
92.471 |
92.636 |
95.470 |
|
Distribution |
|
|
|
|
|
VDss (Log L Kg-1) |
0.194 |
0.173 |
-0.232 |
-0.101 |
|
BBB permeability (log BB) |
-0.644 |
0.034 |
-0.331 |
-0.405 |
|
CNS permeability (log PS) |
-2.244 |
-2..223 |
-1.954 |
-1.943 |
|
Metabolism |
|
|
|
|
|
Substrate CYP |
|
|
|
|
|
2D6 |
No |
No |
No |
No |
|
3A4 |
Yes |
Yes |
Yes |
Yes |
|
Inhibitor |
|
|
|
|
|
2D6 |
No |
No |
No |
No |
|
3A4 |
No |
No |
Yes |
Yes |
|
Excretion |
|
|
|
|
|
Total clearance (Log mL min-1 Kg-1) |
0.266 |
0.328 |
0.426 |
0.538 |
|
Toxicity |
|
|
|
|
|
AMES toxicity |
Yes |
No |
Yes |
Yes |
DISCUSSION
Synthesis of N1-benzoyl pyrazoline compound
The synthesis scheme of N1-benzoyl pyrazoline is presented in Figure 1. The Claisen-Schmidt condensation reaction of 2’-hydroxy acetophenone with aromatic aldehyde derivatives produced chalcones. NaOH is a base catalyst that accelerates this reaction. The chemical structure of chalcone was characterized by FTIR, 1H- dan 13C-NMR. FTIR spectra showed the signal at 1635 (C=O), 1442 (C=C alkene olefin), and 3402 cm-1 (OH). The peaks Hα and Hβ with 15.00 Hz coupling constants represent the chalcone compounds observed at 7.51 and 7.88 ppm, respectively (Bandeira et al., 2019). The 13C-NMR spectra also confirm that Cα dan Cβ peaks were recorded at 118.93 and 147.01 ppm, as well as the carbonyl group, was recorded at 193.77 ppm. This revealed that α,β-unsaturated backbone had been formed, demonstrating a successful synthesis of chalcone derivatives.
Chalcones were reacted further with hydrazine monohydrate by cyclization reaction to obtain N1-hydrogen pyrazoline. The emergence of NH and C=N absorption peaks was found at 3302 and 1597 cm-1, respectively. These two absorptions signals represent the N2 atom and C3 of the pyrazoline ring (Habib et al., 2010). The significant feature of ring pyrazoline was recorded at a chemical shift of 3.10 (dd, 1H, Hα), 3.54 (dd, 1H, Hβ), and 4.83 ppm (t, 1H, Hx) (Wahyuningsih et al., 2019). Meanwhile, the emergence of atoms C4 (-CH2), C5 (-CH-NH), and C3 (-C=N) was found at 41.98, 56.90, and 154.65 ppm, respectively. These results empower the pyrazoline ring produced.
The changing of N1-hydrogen to N1-benzoyl pyrazoline was achieved via an SN2 reaction. N1-hydrogen pyrazoline reacted with benzoyl chloride in the presence of glacial acetic acid as a catalyst. In FTIR spectra, the carbonyl group was recorded at 1620 cm-1, while C=N and OH groups were observed at 1597 and 3448 cm-1, respectively. 1H-NMR spectra revealed structurally N1-benzoyl compound by some aryl proton detected at δ 7.49-7.79 ppm. The significance of the proton pyrazoline ring also appeared on the chemical shift at 3.36, 3.89, and 5.70 ppm for Hα, Hβ, and Hx, respectively. This proton was revealed as a doublet of doublet multiplicity as a result of coupled vicinal of a proton with two methylene protons on C4 position of the pyrazoline ring. Proton Hx of N1-benzoyl pyrazoline was found downfield compared to the Hx proton of N1-hydrogen pyrazoline. Thus, a stronger induction effect ((C=O)N) than the NH group hence proton Hx became deshielded. Meanwhile, 13C-NMR spectra displayed several peaks of aryl ring as well as carbon carbonyl. This result revealed that N1-hydrogen pyrazoline had been converted to N1-benzoyl pyrazoline. The represented peaks of pyrazoline rings such as CH2, CH-N, and C=N were still preserved.
Antibacterial activity
N1-hydrogen and N1-benzoyl pyrazoline were obtained and further evaluated for antibacterial assay to five bacteria. Staphylococcus aureus, Bacillus cereus, and Bacillus subtilis are Gram-positive, while Escherichia coli and Shigella flexneri belong to Gram-negative bacteria. The antibacterial assay result showed that N1-hydrogen pyrazoline exhibited good activity on all bacteria. This is a notion that N1-hydrogen pyrazoline has NH group in pyrazoline ring and hydroxyl (-OH) bearing on ring A as well as ring B contributes to hydrogen interaction with the hydroxyl group of LPSs on Gram-negative. Consequently, cell wall damage occurs (Suryani, 2014). On the other hand, the Gram-positive cell wall contains peptidoglycan so that N1-hydrogen pyrazoline is not able to interfere permeability barrier; as a result, it easily diffuses and impairs the cell wall.
The alteration of N1-hydrogen to N1-benzoyl on the pyrazoline ring decreases activity and tends to favor Gram-positive bacteria. This is due to the benzoyl presence that increases the polarity of the pyrazoline compound; as a consequence, this compound has a strong hydrophilic character. Chauhan et al. (2011) reported that Gram-negative bacteria containing phospholipid in outer lipopolysaccharide (LPSs) would produce impermeable for the drug. Therefore, this reason makes the N1-benzoyl pyrazoline difficult to diffuse into the outer membrane of Gram-negative bacteria. Conversely, N1-benzoyl pyrazoline shows better activity toward Gram-positive. It is supposed to interact well with the hydroxyl group of transpeptidase enzymes such as serin. This interaction results in the interfering synthesized cell wall and causes dramatic lysis of the cell wall. However, this needs further study by molecular modeling to confirm the antibacterial results obtained.
Molecular docking
The antibacterial activities of all compounds are presented in Table 1. The attached benzoyl on N1 position of the pyrazoline ring decreased the antibacterial activity. It was distinct from the preliminary suspicion of this study. On the other hand, molecular docking is a tool used to find binding energy and interaction between ligands with amino acid residue (Saikia and Bordoloi, 2018). All synthetic compounds were docked to ecKAS III enzyme with the 3D protein structure retrieved from the RSCB website (ID PDB: 1hnj). The ecKAS III enzyme is an enzyme that plays a role in fatty acid biosynthesis, the component of membrane cells (Lee et al., 2011). When this enzyme is not working properly to influence the impermeability of membrane cells, bacteria death occurs. The significant role of ecKAS III makes this enzyme a target in new antibiotic discoveries (Nofiana et al., 2019).
All synthetic compounds exhibited strong inhibitory activity toward ecKAS III enzyme via molecular docking study. The binding energy of all compounds spans from -8.1 to -8.3 kcal/mol. Both compounds 3 and 4 gave the lowest binding energy. It means both compounds have a stability docked pose on the active site of the ecKAS III pocket. However, the evaluation of inhibited enzyme activity is not examined only through binding energy. The interaction of synthetic compounds to an amino acid residue is liable to the catalytic activity of the ecKAS III enzyme. The catalytic triad hole of ecKAS III is Cys-His-Asn, the important active side in condensing fatty acid process (Lee et al., 2011). The blocked of these amino acids could be the proposed mode mechanism of disruption of membrane cells.
The presence of hydroxyl group on ring B of pyrazoline contributes to the hydrogen bonding with Asn247. This amino acid is located at the active site of the ecKAS III pocket (Figure 3a). This confirms that four synthetic compounds give a satisfactory result. According to the antibacterial outcome (Table 1), N1-hydrogen pyrazoline derivatives demonstrate good inhibitory to Gram-positive and Gram-negative bacteria. This compound could bind both Asn247 and Cys112. They are important amino acids inside the catalytic of the ecKAS III enzyme. In addition, the type of interaction provides supporting information on how compound 1 has slightly better energy binding than compound 2. Compound 1 can form a hydrogen bond that obtains a stronger bond than the hydrophobic of compound 2.
To identify the impact of the addition of benzoyl on the N1 position of the pyrazoline ring, we found that benzoyl removed the Cys112 interaction. Also, benzoyl enhances steric hindrance; compounds 3 and 4 could not establish the interaction of either hydrogen or hydrophobic bonding toward Cys112, likewise on compounds 1 and 2. In brief, the suggested modification of the pyrazoline compound is on rings A and B, but it preserves the pyrazoline ring originally to earn preferable inhibitory activity.
ADMET study
All compound obtained was further analyzed through ADMET in silico study by pkCSM tool. As shown in Table 3, compounds have good absorption capacity in the body. They have an intestinal absorption percentage value of up to 90%, which means compounds have excellent absorption. The distributed ability at a total dose in blood plasma is represented by the steady-state volume of distribution (VDss). The lowest distribution is considered by VDss value about < - 0.15 and good if above 0.45. Based on the analysis, all compounds have VDss value in the range that revealed they distribute equally in blood plasma except compound 3. BBB permeability is the value that describes the capacity of a drug across the brain barrier membrane, while CNS permeability depicts the ability of a drug to penetrate the central nervous system. The result analysis showed that compounds across the CNS barrier in moderate to good (high, log PS > -2; low, log PS < -3). Compound 2 only passes the brain barrier membrane while the other compound has poorly across it (good penetrate, log BB > 0.3; poor penetrate, log BB < -1) (Pires et al., 2015).
Drug metabolism describes the drug degraded by the cytochrome P450 enzyme to produce harmless metabolite and eliminate it. The cytochrome P450 has played a role in the metabolism of drugs at phase I through oxidation reaction (Hadni and Elhallaoui, 2020). In this present study, the used isoforms were CYPD26 and CYP3A4. Both enzymes play a role in drug metabolism and xenobiotic (Arici and Özhan, 2017; Basheer and Kerem, 2015). Table 6 shows that all compounds are not acting as an inhibitor to both enzymes, while compounds 3 and 4 only inhibit the CYP3A4 enzyme. This is a notion that synthetic compound has good metabolism with P450 cytochrome.
The character of drug excretion is represented as the total clearance value. This value describes how a drug compound could be eliminated. The lowest total clearance means the drug has persistence over the elimination process (Hadni and Elhallaoui, 2020). The whole synthetic compound has a high total clearance value. Thereby, they are easily breaking down in the elimination process. Afterward, the toxicity of synthetic compounds is also interesting to study in drug development. In most cases, plenty of drug compound has good biological activity but toxic character. In general, the ADMET result informs that compound 2 has the potential to be further developed as a drug and is non-toxic.
CONCLUSION
N1-hydrogen and N1-benzoyl pyrazoline have been successfully synthesized, and their structures were confirmed by FTIR, 1H- and 13C-NMR spectra. N1-hydrogen pyrazoline was a compromising compound as an antibacterial agent through in vitro and in silico studies from molecular docking and ADMET analysis. The presence of benzoyl on N1 in the pyrazoline ring decreased the inhibition zone and was active on Gram-positive bacteria only. Compound 2 has no toxic properties and showed good ADMET character from the in silico study.
AUTHOR CONTRIBUTIONS
Tutik Dwi Wahyuningsih conceived the design research, supported the analysis and research material, and revised the manuscript. Putra Jiwamurwa Pama Tjitda conducted the experiment, drafted and revised the manuscript. Jumina supervised the research. The final manuscript has been reviewed and approved by all authors.
CONFLICT OF INTEREST
There is no conflict of interest between any authors in this research.
REFERENCES
Abeed, A.A.O., Jaleel, G.A.A., and Youssef, M.S.K. 2019. Novel heterocyclic hybrids based on 2-pyrazoline: Synthesis and assessment of anti-inflammatory and analgesic activities. Current Organic Synthesis. 16: 921–930.
Alkamaly, O.M., Altwaijry, N., Sabour, R., and Harras, M.F. 2021. Dual EGFR/VEGFR2 inhibitors and apoptosis inducers: Synthesis and antitumor activity of novel pyrazoline derivatives. Archiv der Pharmazie. 354: 1–17.
Arici, M. and Özhan, G. 2017. CYP2C9, CYPC19 and CYP2D6 Gene profiles and gene susceptibility to drug response and toxicity in Turkish population. Saudi Pharmaceutical Journal. 25: 376–380.
Azarifar, D., and Shaebanzadeh, M. 2002. Synthesis and characterization of new 3,5-Dinaphthyl substituted 2-Pyrazolines and study of their antimicrobial activity. Molecules. 7: 885–895.
Bandeira, P.N., Lemos, T.L.G., Santos, H.S., de Carvalho, M.C.S., Pinheiro, D.P., de Moraes Filho, M.O., Pessoa, C., Barros-Nepomuceno, F.W.A., Rodrigues, T.H.S., Ribeiro, P.R.V., Magalhães, H.S., and Teixeira, A.M.R. 2019. Synthesis, structural characterization, and cytotoxic evaluation of chalcone derivatives. Medicinal Chemistry Research. 28: 2037–2049.
Basheer, L., and Kerem, Z. 2015. Interactions between CYP3A4 and dietary polyphenols. Oxidative Medicine and Cellular Longevity. 2015. 1-15.
Chauhan, A., Sharma, P.K., Kaushik, N., and Kumar, N. 2011. Synthesis of novel pyrazole analogues as efficacious antimicrobial agents. International Journal of Pharmacy and Pharmaceutical Sciences. 3: 166–176.
Da, S.C.J., Da, S.R.R., Da, S.L.C.K., Do, S.B.B.D., De, P.D.S.C.H.T., Ferreira, E.F.B., Dos, S.B.R., Campos, J.M., Da, C.M.W.J., Dos, S.C.B.R. 2018. An in silico study of the antioxidant ability for two caffeine analogs using molecular docking and quantum chemical methods. Molecules. 23: 1-17.
De, D., and Saha, S. 2019. Synthesis and characterisation of 1,3,5 triphenyl-2-pyrazoline for antimicrobial activity. World Journal of Pharmaceutical Research. 8: 883–897.
Desai, N.C., Pandya, D., and Vaja, D. 2017. Synthesis and antimicrobial activity of some heterocyclic compounds bearing benzimidazole and pyrazoline motifs. Medicinal Chemistry Research. 27: 52–60.
Fernandes, J., Revanasiddappa, B.C., Ishwarbhat, K., Vijay Kumar, M., D’Souza, L., and Alva, S.S. 2017. Synthesis and in-vitro anti-inflammatory activity of novel pyrazoline derivatives. Research Journal of Pharmacy and Technology. 10: 1679–1682.
Freitas, T.S.D., Xavier, J.D.C., Pereira, R.L.S., Rocha, J.E., Muniz, D.F., Da Silva, P.T., Da Hora, J.P., Dos Santos, H.S., Bandeira, P.N., Nogueira, C.E.S., Teixeira, A.M.R., and Coutinho, H.D.M. 2020. Direct antibacterial and antibiotic resistance modulatory activity of chalcones synthesized from the natural product 2-Hydroxy-3,4,6-trimethoxyacetophenone. FEMS Microbiology Letters. 367: 1–8.
Habib, S.I., Kulkarni, P.A., and Konda, S.G. 2010. A convenient synthesis of some novel hydroxy pyrazolines under thermal condition and their antimicrobial activity. Asian Journal of Research in Chemistry. 2: 1960–1965.
Hadni, H., and Elhallaoui, M. 2020. 2D and 3D-QSAR, molecular docking and ADMET properties: In silico studies of azaaurones as antimalarial agents. New Journal of Chemistry. 44: 6553–6565.
Hoz, A.D.L., Alkorta, I., and Elguero, J. 2021. The mechanism of the reaction of hydrazines with α,β-unsaturated carbonyl compounds to afford hydrazones and 2-pyrazolines (4,5-dihydro-1H-pyrazoles): Experimental and theoretical results. Tetrahedron. 97: 1–19.
Illicachi, L.A., Montalvo-Acosta, J.J., Insuasty, A., Quiroga, J., Abonia, R., Sortino, M., Zacchino, S., and Insuasty, B. 2017. Synthesis and DFT calculations of novel vanillin-chalcones and their 3-aryl-5-(4-(2-(dimethylamino)- ethoxy)-3-methoxyphenyl)-4,5-dihydro-1H-pyrazole-1-carbaldehyde derivatives as antifungal agents. Molecules. 22: 1-20.
Jadhav, S.A., Shirote, P.J., Kulkarni, K.M., and Patil, V.M. 2018. Synthesis, spectral analysis and anticancer evaluation of novel pyrazoline derivatives. American Journal of PharmTech Research. 8:303–315.
Kumar, A., Varadaraj, B.G., and Singla, R.K. 2013. Synthesis and evaluation of antioxidant activity of novel 3,5-Disubstituted-2-Pyrazolines. Bulletin of Faculty of Pharmacy. 51: 167–173.
Lee, J.Y., Lee, E., Jeong, K.W., and Kim, Y. 2011. Antimicrobial flavonoid, 3,6-dihydroxyflavone, have dual inhibitory activity against KAS III and KAS I. Bulletin of the Korean Chemical Society. 32: 3219–3222.
Li, B., and Webster, T.J. 2018. Bacteria antibiotic resistance: New challenges and opportunities for implant-associated orthopedic infections. Journal of Orthopaedic Research. 36: 22–32.
Nofiana, R., Philmus, B., Nindita, Y., and Mahmud, T. 2019. 3-Ketoacyl-ACP synthase (KAS) III homologues and their roles in natural product biosynthesis. Medicinal Chemistry Communications. 10: 1517–1530.
Patel, A.R., Badmanaban, R., Sen, D.J., and Patel, C.N. 2013. Design, synthesis and antimicrobial, antifungal and anti-inflammatory evaluation of some (4-substituted phenyl)[5-(4-substituted phenyl)-3-phenyl- 4,5-dihydro-1H-pyrazol-1-yl]-methanone derivatives. American Journal of Advanced Drug Delivery. 2: 113–127.
Pires, D.E.V., Blundell, T.L., and Ascher, D.B. 2015. pkCSM: Predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. Journal of Medicinal Chemistry. 58: 4066–4072.
Pola, S., Banoth, K.K., Sankaranarayanan, M., Ummani, R., and Garlapati, A. 2020. Design, synthesis, in silico studies, and evaluation of novel chalcones and their pyrazoline derivatives for antibacterial and antitubercular activities. Medicinal Chemistry Research. 29: 1819–1835.
Qiu, X., Janson, C.A., Smith, W.W., Head, M., Lonsdale, J., and Konstantinidis, A.K. 2001. Refined structures of β-ketoacyl-acyl carrier protein synthase III. Journal of Molecular Biology. 307: 341–356.
Rana, M., Arif, R., Khan, F.I., Maurya, V., Singh, R., Faizan, M.I., Yasmeen, S., Dar, S.H., Alam, R., Sahu, A., Ahmad, T., and Rahisuddin. 2021. Pyrazoline analogs as potential anticancer agents and their apoptosis, molecular docking, MD simulation, DNA binding and antioxidant studies. Bioorganic Chemistry. 108: 1-17.
Saikia, S., and Bordoloi, M. 2018. Molecular docking: Challenges, advances and its use in drug discovery perspective. Current Drug Targets. 20: 501–521.
Setyawati, A., Wahyuningsih, T.D., and Purwono, B. 2017. Syntheses of novel pyrazolines as antibacterial agents from natural product vanillin. Asian Journal of Chemistry. 29: 454–456.
Suma, A.A., Wahyuningsih, T.D., and Pranowo, D. 2017. Synthesis and antibacterial activities of N-phenylpyrazolines from veratraldehyde. Material Science Forum. 901: 124–132.
Suryani, O. 2014. Sintesis dan Uji Aktivitas Antibakteri Secara In Vitro Senyawa Turunan N-Asetil-2-Pirazolina Tersubstitusi Gugus Hidroksi Berbahan Dasar Vanilin. Universitas Gadjah Mada, Yogyakarta.
Trott, O., and Olson, A.J. 2009. Software news and update AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry. 31: 454–461.
Wahyuningsih, T.D., Suma, A.A.T., and Astuti, E. 2019. Synthesis, anticancer activity, and docking study of N-acetyl pyrazolines from veratraldehyde. Journal of Applied Pharmaceutical Science. 9: 14–20.
OPEN access freely available online
Chiang Mai University Journal of Natural Sciences [ISSN 16851994]
Chiang Mai University, Thailand.
https://cmuj.cmu.ac.th
Putra Jiwamurwa Pama Tjitda1, Jumina and Tutik Dwi Wahyuningsih2, *
1 Department of Pharmacy, Health Polytechnic of Kupang, Kupang 85111, Indonesia.
2 Department Chemistry, Faculty of Mathematics and Natural Sciences, Universitas Gadjah Mada, Yogyakarta 55281, Indonesia
Corresponding author: Tutik Dwi Wahyuningsih, E-mail: tutikdw@ugm.ac.id
Total Article Views
Editor: Wipawadee Yooin,
Chiang Mai University, Thailand
Article history:
Received: March 3, 2022;
Revised: June 16, 2022;
Accepted: June 17, 2022;
Published online:June 21, 2022

