
Host Range of Listeria Prophages Induced from Lysogenic Listeria Isolates from Foods and Food-related Environments in Thailand. Hue Thi Kim Vu, Soottawat Benjakul, Varaporn Vuddhakul, and Kitiya Vongkamjan*
Published Date : 2019-04-1
DOI : https://doi.org/10.12982/CMUJNS.2019.0011
Journal Issues :
Number 2 , April-June 2019
ABSTRACT
Prophages have been shown to be associated with improving the survival and fitness of Listeria spp., especially Listeria monocytogenes-a fatal foodborne pathogen. Bioinformatics has revealed the presence of many prophages in Listeria genomes. However, understanding the distribution of prophages in Listeria isolates is limited to those that have no sequences available on the database. In this study, Mitomycin C induction was used to obtain prophages in free-floating form. Among 236 isolates from various sources, prophages were induced from 13/108 (12%) isolates of L. monocytogenes and 10/128 (7.8%) isolates of Listeria spp. Of 39 induced phages obtained, most phages were originated from foods. Phenotypic characterization was performed by the host range determination against 17 hosts representing 9 major serotypes of L. monocytogenes and 4 other species of Listeria. Induced phages were classified into three groups. The majority of phages (groups A and C) were host-specific- phages with the ability to lyse one to seven (<42%) hosts. However, five phages (group B) showed broader host range than phages in other groups, which could lyse 8–10 (47–59%) hosts. Both induced and isolated Listeria phages showed a high ability to lyse strains of L.monocytogenes serotype 4, while the induced phages showed narrow host range compared to the isolated phages. Host range data allows the prediction of particular L. monocytogenes subtypes or Listeria species that could be affected by the induced phages, leading to gene transfer upon phage infection. Information obtained here is useful to understand the diversity and role of prophages in Listeria genomes.
Keywords: Listeria spp., Listeria prophage, Food processing, Phage host range
INTRODUCTION
Listeria monocytogenes is an important foodborne pathogen causing potentially fatal listeriosis infections with a high mortality rate of up to 30% (Swaminathan and Gerner-Smidt, 2007). The genus of Listeria comprises 17 species, including two pathogenic species L. monocytogenes and L. ivanovii. Previous studies reported that various types of foods could be contaminated with L. monocytogenes (Vongkamjan et al., 2015, 2016). This pathogen has been shown the ability to be well adapted and survive longer in specific environments (Palumbo and Williams, 1991; Tolvanen et al., 2008; Burgess et al., 2016). Prophages have been shown the linkage on providing greater survival and fitness of Listeria spp., especially L. monocytogenes (Orsi et al., 2008; Verghese et al., 2011).
Prophage is a phage-related sequence which has integrated into bacterial chromosome and become part of the bacterial genome. Prophages have been reported to be commonly present in the Listeria genomes (Nelson et al., 2004; Kuenne et al., 2013), for example, L. monocytogenes strains F6854, L99, HCC23, J0161 (Kuenne et al., 2013; den Bakker et al., 2013). L. innocua strain CLIP11262 harbored up to six prophage (-like) elements, including 5 prophages and 1 monocin (Nelson et al., 2004). Previous studies mostly applied bioinformatics analysis to search for prophage regions in the genome of Listeria. This approach restricts information on prophage diversity to those lysogens (prophage-carrying Listeria) that have no sequences available on the database.
Induction is a mechanism by which prophage can be induced to escape a dying host and enter the lytic cycle, producing a free-floating form called induced phage. In previous studies, the occurrence of prophages has been measured by induction from the lysogenic isolates (Jiang and Paul, 1996; Chen et al., 2006). Some antibiotics, UV radiation, sunlight, temperature and pressure have been previously reported as common inducing-agents for prophage induction (Jiang and Paul, 1996; López et al., 2014). However, mitomycin C has been regarded as the most popular agent to induce prophages and produce infective phages (Loessner et al., 1991; Verghese et al., 2011).
Phage characterization is useful for obtaining information on the diversity of both phages isolated from natural sources (freely isolated phages) and phages induced from the lysogens. Previous studies have performed a phenotypic characterization by host range determination in Listeria phages isolated from natural dairy farms (Vongkamjan et al., 2012) or seafood processing (Vongkamjan et al., 2017) as well as turkey processing plant environments (Kim et al., 2008). However, there is still limited information on the characterization of Listeria phages induced from lysogenic isolates.
In this study, mitomycin C induction was performed to examine the prevalence of prophage-carrying Listeria among isolates obtained from various foods and food-related environments. The newly induced Listeria phages were then characterized phenotypically by host range determination and compared to that of freely isolated Listeria phages from seafood environment samples. This information is useful to link or speculate particular sources that showed high prevalence and diversity of prophages, which is a potential factor for the bacteria to gain better survival and present in foods and food-related environments.
MATERIALS AND METHODS
L. monocytogenes and Listeria spp. used in this study
Atotal of 236 Listeria isolates consisting of 108 isolates of L. monocytogenes and 128 isolates of Listeria spp. (non-monocytogenes) (Table 1) were tested for the presence of prophage by mitomycin C induction. These isolates were previously obtained by the standard Listeria isolation protocols (Bacteriological Analytical Manual of the US Food and Drug Administration) and confirmed by PCR (Vongkamjan et al., 2015, 2016, 2017). Listeria isolates were classified into five categories based on the sources (foods and food-related environments). The food sources included raw and ready-to-eat products of animal origin (44 isolates), seafood/ aquatic origin (103 isolates) or vegetable origin (8 isolates). The food- related environments included food contact surfaces (41 isolates), for example, buckets/ trays/ baskets used to contain products, blenders, knives, gloves, tables; and non-food contact surfaces (40 isolates) such as digital scales, cleaning sponges, and drains.
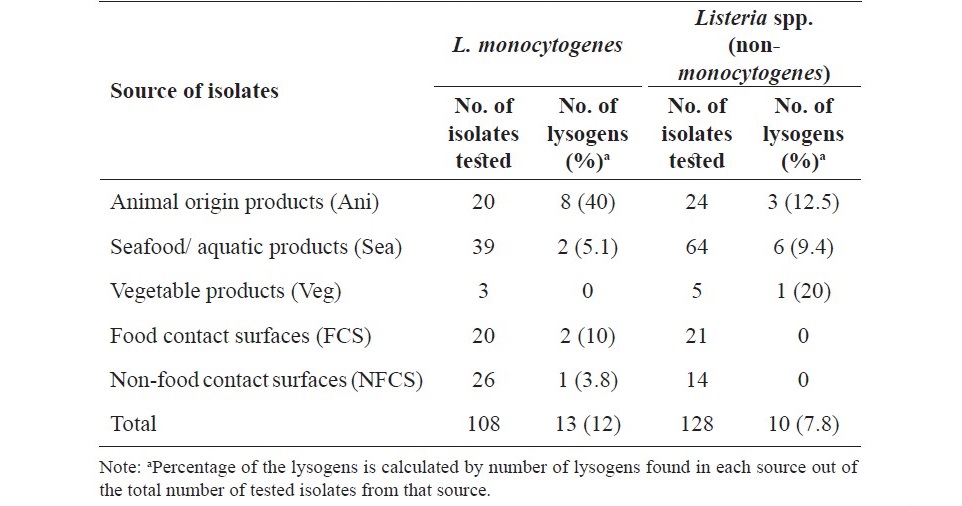
Table 1. Prevalence of Listeria lysogens among the isolates of L. monocytogenes and Listeria spp. from various sources.
Four L. monocytogenes reference strains (Mack, FSL F2-695, FSL J1-208 and F2365 were obtained from Food Safety Lab (FSL), Cornell University) were used as propagating hosts for phage induction, purification and lysate preparation. Host range determination was performed using the same panel of hosts as previously studied by Vongkamjan et al. (2017). In which, 13 strains of L. monocytogenes representing nine major serotypes of L. monocytogenes included serotypes 4a, 4b, 4c (n=7), serotypes 1/2a, 1/2b, 1/2c (n=3) and serotypes 3a, 3b, 3c (n=3). Additional four strains representing other four species (L. innocua, L. ivanovii, L. marthii, L. seeligeri) were also included. All of Listeria strains/ isolates were kept in Brain Heart Infusion (BHI, Oxoid, UK) broth with 15% glycerol and stored at -80°C at the department of Food Technology, Prince of Songkla University (PSU), Thailand.
Induction of Listeria prophages by mitomycin C
Cultures of Listeria isolates were prepared by inoculating a single colony of each tested isolate in 5 ml of Luria Bertani (LB, Himedia, Mumbai, India) broth with a–supplement of 50 mM morpholinepropanesulfonic acid (MOPS), 1% (wt/ vol) glucose, 10 mM CaCl2, and 10 mM MgCl2 (LB-MOPS-Glu-salts) (Vongkamjan et al., 2012). The tested isolate was incubated at 30°C with shaking (220 rpm) for 8–10 h to reach a 600 nm optical density (OD600) of 0.4 to 0.5. The culture of each tested isolate was mixed with mitomycin C (Sigma-Aldrich, St Louis, USA)
to a final concentration of 1 µg/ml, followed by additional incubation for 7 h at the same conditions (modified from Fortier and Moineau, 2007). A quantity of 200 µl of mitomycin C-treated culture was mixed with 100 µl of each propagating host overnight culture in 2 ml of LB-MOPS-Glu-salts broth. Then, the co-culture was incubated for 18 h at 30°C with shaking (220 rpm). To isolate the induced phage, 1 ml of overnight co-culture was centrifuged at 10,000×g for 15 min at 4°C. The supernatant was used to prepare an overlay using 0.7% LB-MOPS top- agar with a 10-fold-diluted overnight culture of a given propagating host, then poured on the 1.5% LB-bottom-agar (Vongkamjan et al., 2012). After an overnight incubation at 30°C, each plate was examined for plaque-forming on the host lawn. The presence of plaque formation indicated the appearance of induced phage, the tested Listeria isolate was recorded as a lysogen. Each different types of plaque morphology observed (differentiated by a plaque diameter between 0.5–2mm, star/ round shape, turbid/ translucent/clear zone) was recorded as a single induced phage.
Purification of induced Listeria phages and preparation of phage lysates
An isolated plaque of each plaque-morphology type was selected for phage purification using the double layer method. This step was conducted three times to ensure a pure phage following the protocol previously described by Vongkamjan et al. (2012). Upon the third purification, a single plaque was used to prepare a high titer phage stock using two confluent lysis plates. A volume of 7 ml of Phosphate Buffer Saline (PBS) with a pH of 7.4 was added to each plate, followed by a centrifugation at 5,000×g for 15 min at 4°C and filtration through a 0.22-µm pore- size filter. Phage titers were determined by spotting 5-µl of eight of 10-fold serial phage lysate dilutions on the prepared lawn of the propagating host, as mentioned above.
Determination of phage host range
The host range of each induced Listeria phage was determined with the same host panel as previously used to characterize the host range of Listeria phages isolated from seafood processing samples (Vongkamjan et al., 2017). A quantity of 5-µl of the diluted phage representing 100×RTD (routine test dilution) (approximately 106–107 PFU/ml) was spotted on the prepared lawn of each host, followed by an overnight incubation at 30°C. Each spot on the lawn was examined and recorded as lysing (+), which known as the presence of multiple or single plaque(s), turbid or confluent lysis at the spotting area or no lysing (-). The experiment was carried out in triplicate. The induced phage was considered that could lyse a tested host when plaque forming was observed in at least two replicates. Lysis ability of the induced phages against the tested hosts was used to classify these phages into clusters. Hierarchical clustering was performed using the R program version 3.1.2. with Ward’s method and binary distance as previously described (Vongkamjan et al., 2012).
Comparison of host ranges between newly induced Listeria phages and freely isolated Listeria phages from seafood processing environments in Thailand
Host range data of 39 newly induced Listeria phages in this study and the data of 29 freely isolated Listeria phages from the previous study by Vongkamjan et al. (2017) were compared. These two sets of Listeria phages were tested against the same panel of reference hosts in order to examine the difference between lysis abilities of induced phages and isolated phages. A previously reported Listeria phage LP124 was used as a control for both sets of induced and isolated Listeria phages.
RESULTS
Occurrence of Listeria lysogens among the Listeria isolates from various sources
From total of 236 Listeria isolates used for determination the presence of prophages by mitomycin C induction, 23 Listeria isolates were lysogens (Table 1). Of these, 13/108 (12%) lysogens were L. monocytogenes and 10/128 (7.8%) lysogens were Listeria spp. Among the 108 tested L. monocytogenes isolates, the prevalence of lysogens was highest in the isolates from Ani (40%) followed by FCS (10%) and Sea (5.1%). Among the 128 Listeria spp. isolates, prevalence of lysogens was highest in the isolates from Veg (20%), followed by Ani (12.5%) and Sea (9.4%). In particular, none of L. monocytogenes isolates from Veg or Listeria spp. isolates from FCS and NFCS were lysogens.
Distribution of prophages in Listeria lysogens from various sources
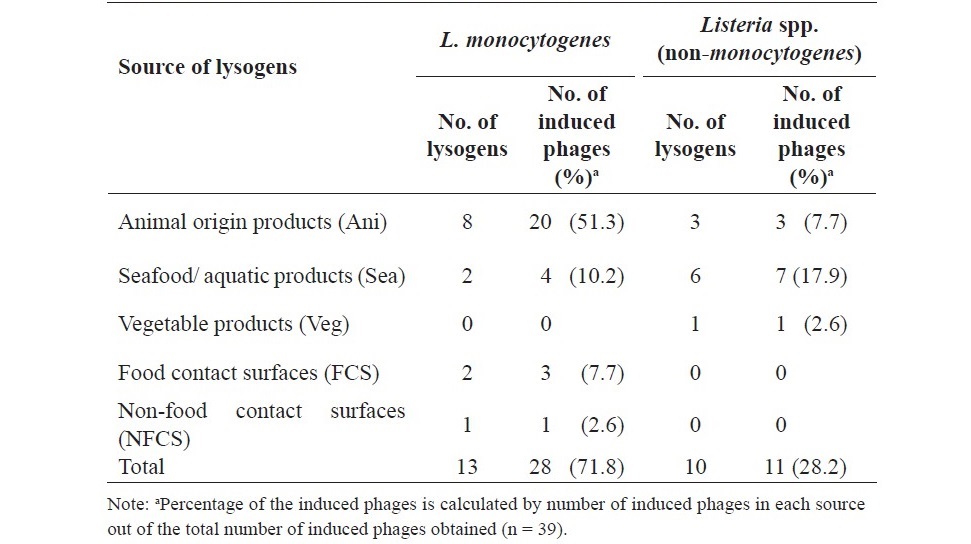
Based on the presence of distinct types of plaque morphology observed, 39 induced Listeria phages were obtained from 23 lysogens (Table 2). From the 13 lysogens of L. monocytogenes, 28 induced Listeria phages were obtained upon mitomycin C induction. Of these, 20 phages (51.3%) were from L. monocytogenes lysogens of Ani, the remaining phages was from L. monocytogenes lysogens of Sea (10.2%), FCS (7.7%) and NFCS (2.6%). From the 10 lysogens of Listeria spp., 11 induced phages were obtained. All of which were induced from Listeria spp. lysogens of the food sources with Sea (17.9%), followed by Ani (7.7%) and Veg (2.6%).
Table 2. Distribution of prophages in Listeria lysogens from various sources.
Phages induced from both L. monocytogenes lysogens and Listeria spp. (non-monocytogenes) lysogens were able to be propagated in L. monocytogenes hosts. By using different serotypes of L. monocytogenes as propagating hosts, distribution of induced Listeria phages was various with between 6 and 12 induced phages obtained in each propagating host (data not shown).
Lysis ability of induced Listeria phages on Listeria hosts
All 39 induced Listeria phages were used for the host range determination against 17 reference strains representing nine serotypes of L. monocytogenes and four other Listeria spp. (Figure 1). Clustering analysis using the R program classified these induced phages into 22 lysis profiles, representing three host range groups, labeled A to C. Group C contained 17 host-specific-phages, which could lyse only one to three (<20%) hosts of L. monocytogenes serotype 4. Group A contained 17 phages with similar host range as those in group C (host-specific phages), but they could lyse more hosts of L. monocytogenes serotype 4 and L. marthii. However, the remaining five phages (group B) showed broader host range than phages in group A and C. In which, group B phages could lyse 8–10 (47–59%) hosts tested.

Figure 1. Clustering analysis of 39 induced Listeria phages against 17 Listeria hosts using the R program version 3.1.2. X-axis represents 13 L. monocytogenes and 4 Listeria spp. host strains. Y-axis represents 39 induced Listeria phages. Black represents no-lysing and light-gray represents lysing on a given host strain. Four references strains marked with “*” were used as the propagating hosts for prophage induction, phage purification and phage lysate preparation.
Of the tested hosts, L. monocytogenes serotype 4 strains were highly susceptible to the induced phages, which were lysed by up to 30/39 phages tested. For example, strain F2365 (serotype 4b) was lysed by 30 induced phages, strain FSL J1-208 (serotype 4a) was lysed by 24 induced phages. In addition, L. marthii (FSL C7-0084) was lysed by 19 induced phages. In contrast, five strains of L. monocytogenes serotypes 1/2 (1/2b, 1/2c) and serotypes 3 (3a, 3b, 3c) and one strain of L. seeligeri shown to be resistant to all of the induced phages.
Comparison of host ranges between induced Listeria phages and Listeria phages freely isolated in Thailand
By having the same set of Listeria hosts used in the phage host range determination, phage lysis profiles of our induced phages were compared to those of freely isolated Listeria phages from a previous study (Vongkamjan et al., 2017). Figure 2 showed the percentage of induced/isolated phages which were able to infect each host. In general, most of the induced phages were host-specific with the ability of lysing < 42% of the hosts, while the freely isolated phages presented both broad and narrow host ranges with the ability of lysing > 50% of the hosts. For instance, the lysis ability of isolated phages in each host was at least double that of the induced phages. All of the isolated phages could lyse serotype 4 strains (except F2365) and L. marthii. However, all of the induced phages were resistant to most hosts representing L. monocytogenes serotype 1/2 (except Mack), serotype 3, L. innocua and L. seeligeri. Interestingly, none of phages in both groups of induced and isolated phages could lyse two hosts of L.momocytogenes serotype 3b and serotype 3c.
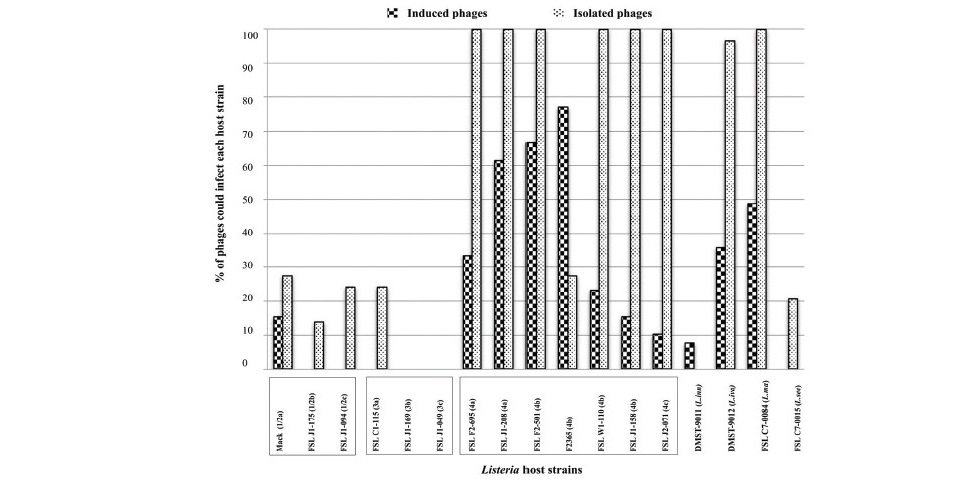
Figure 2. Lysis ability of the induced Listeria phages (n=39) and freely isolated Listeria phages (n=29) against Listeria hosts. X-axis represents 13 L. monocytogenes and 4 Listeria spp. host strains. Y-axis represents percentage of induced Listeria phages/isolated Listeria phages which could lyse each host.
DISCUSSION
High distribution of prophages in Listeria isolates from foods, especially from animal origin products
Listeria lysogens are known to be widespread among Listeria spp. (Loessner et al., 1991) and the induction of prophages by mitomycin C was carried out to examine the prophage prevalence. Among 236 Listeria isolates, 23 isolates (9.7%) were lysogens and produced plaque in L. monocytogenes propagating host(s). Listeria isolates from food sources (Ani, Sea and Veg) were more likely to carry inducible phages (12.9%) than those from non-food sources (3.7%). Of the food source, up to 51.3% L. monocytogenes isolates of animal origin products were lysogens. Prevalence of prophages found here were similar to the report in a previous study (Ferreira et al., 2011), in which prophages were found in 53.7% of L. monocytogenes isolates (n=41) from meat processing plants. Overall, findings suggest that Listeria isolates of animal origin or related environments are more likely to carry prophages.
Although reports relating to Listeria prophages are limited, in other bacteria such as Staphylococcus aureus, phages were obtained from meat (30%), seafood (67%) and dairy-related environments (97%), (Gutiérrez et al., 2016). Another study reported that none of the nine isolates of Clostridium difficile from meat carried prophages, nine out of 49 isolates of C. difficile from animal and human origins carried prophages (Sekulovic et al., 2014).
Induced Listeria phages appear to be host-specific with a higher ability to lyse L. monocytogenes serotype 4 than other serotypes
Majority of induced Listeria phages (87%) belonged to host range groups A and C which represented as host-specific phages. Similar findings have previously been reported that induced phages showed narrow host range (Horgan et al., 2010; Wright et al., 2013). The host range data pointed out that both sets of phages could lyse most strains of L. monocytogenes serotype 4 and L. marthii. Similar findings were also observed in the isolated Listeria phages from silage (Vongkamjan et al., 2012). In addition, another study reported that Listeria phages isolated from a turkey processing environment could lyse most strains of L. monocytogenes serotype 1/2a and serotype 4b (Kim et al., 2008). Finding suggests that not only the isolated Listeria phages, but also the induced Listeria phages showed a high ability in lysing L. monocytogenes serotype 4. One possible explanation for the difference of phage susceptibility between serotypes of L. monocytogenes is the differences in the structures of wall teichoic acids (WTA). It has been reported that L. monocytogenes of serotype 4 contain a WTA structure with terminal glucose and galactose residues, which are reported to be important for phage adsorption (Wendlinger et al., 1996; Eugster and Loessner, 2011).
Most of the induced Listeria phages could not lyse strains of serotype 1/2 (1/2b, 1/2c) and serotype 3 (3a, 3b, 3c), L. innocua and L. seeligeri. It is of interest that serotype 1/2a is linked to the increased cases of listeriosis in the last decade (Mammina et al., 2013; Marini et al., 2016). This suggests the possible relationship between resistance to phage infection and higher survival and virulence of this serotype. Alternative explanation is that these hosts may harbor certain phage-related sequences to aid their survival in the environment without being affected by phage lysis. As explained in a previous study, Oenococcus oeni isolates that were phage-resistant contained a sequence that was homologous to the tested phage sequence (Poblet-Icart et al., 1998). Our understanding is still limited to clarify the resistance mechanism in these host strains. Further experiment, for example, prophage induction of these Listeria hosts and genome sequencing analysis are needed to elucidate this phenomenon.
Induced Listeria phages show a narrow host range as compared to freely isolated Listeria phages
Overall, induced Listeria phages showed narrow host range as compared to the isolated Listeria phages. Previous studies have been reported that the isolated Listeria phages from turkey processing showed a wider host range (Kim et al., 2008), from silage at dairy farm showed both narrow and broad host range (Vongkamjan et al., 2012). This can be explained by the unique characteristic of induced phage as it has the ability to incorporate its genome into the host chromosome instead of only lysing the host. In previous studies, for example, Loessner and Busse (1990) also reported that the phages isolated from lysogenic strains had a narrower lytic spectrum than that of lytic phages. The host range data suggests that not only induced phages but also isolated phages were less susceptible to strains of L. monocytogenes serotype 1/2 and serotype 3 rather than serotype 4 strains.
CONCLUSION
In summary, this study provides new insights into the phenotypic characteristics of Listeria phages induced from Listeria isolates from various food and food-related environments. Prophage distribution data could be linked to particular sources where Listeria prophage-carrying isolates are prevalent. These isolates of L. monocytogenes or other Listeria spp. may gain the ability to better survive in foods and processing environments. The newly induced Listeria phages described herein provide a baseline for further study on prophage diversity and the role of prophages in facilitating the survival and fitness of Listeria hosts in foods and food-related environments.
ACKNOWLEDGEMENTS
This project was financially supported by Prince of Songkla University (AGR600584S; to KV). Funding from the TRF Distinguished Research Professor Grant (to SB) and the Higher Education Research Promotion and the Project Office of the Higher Education Commission (to HTKV) is also acknowledged.
REFERENCES
den Bakker, H.C., Desjardins, C.A., Griggs, A.D., Peters, J.E., Zeng, Q., Young, S.K., Kodira, C.D., Yandava, C., Hepburn, T.A., Haas, B.J., et al. 2013. Evolutionary dynamics of the accessory genome of Listeria monocytogenes. PLoS ONE. 8: e67511. https://doi.org/10.1371/journal.pone.0067511
Burgess, C.M., Gianotti, A., Gruzdev, N., Holah, J., Knøchel, S., Lehner, A., Margas, E., Esser, S. S., Sela, S., and Tresse, O. 2016. The response of foodborne pathogens to osmotic and desiccation stresses in the food chain. International Journal of Food Microbiology. 221: 37–53. https://doi.org/10.1016/j.ijfood micro.2015.12.014
Chen, F., Wang, K., Stewart, J., and Belas, R. 2006. Induction of multiple prophages from a marine bacterium: a genomic approach. Applied and Environmental Microbiology. 72: 4995–5001. https://doi.org/10.1128/AEM.00056-06
Eugster, M.R., and Loessner, M.J. 2011. Rapid analysis of Listeria monocytogenes cell wall teichoic acid carbohydrates by ESI-MS/MS. PLoS ONE. 6: e21500. https://doi.org/10.1371/journal.pone.0021500
Ferreira, V., Barbosa, J., Stasiewicz, M., Vongkamjan, K., Moreno Switt, A., Hogg, T., Gibbs, P., Teixeira, P., and Wiedmann, M. 2011. Diverse geno- and phenotypes of persistent Listeria monocytogenes isolates from fermented meat sausage production facilities in Portugal. Applied and Environmental Microbiology. 77: 2701–2715. https://doi.org/10.1128/AEM.02553-10
Fortier, L.C., and Moineau, S. 2007. Morphological and genetic diversity of temperate phages in Clostridium difficile. Applied and Environmental Microbiology. 73: 7358–7366. https://doi.org/10.1128/AEM.00582-07
Gutiérrez, D., Rodríguez-Rubio, L., García, P., Billington, C., Premarante, A., Rodríguez, A., and Martínez, B. 2016. Phage sensitivity and prophage carriage in Staphylococcus aureus isolated from foods in Spain and New Zealand. International Journal of Food Microbiology. 230: 16–20. https://doi.org/10.1016/j.ijfoodmicro.2016.04.019
Horgan, M., O’Sullivan, O., Coffey, A., Fitzgerald, G.F., van Sinderen, D., McAuliffe, O., and Ross, R.P. 2010. Genome analysis of the Clostridium difficile phage ΦCD6356, a temperate phage of the Siphoviridae family. Gene. 462: 34–43. https://doi.org/10.1016/j.gene.2010.04.010
Jiang, S., and Paul, J. 1996. Occurrence of lysogenic bacteria in marine microbial communities as determined by prophage induction. Marine Ecology Progress Series. 142: 27–38.
Kim, J.W., Siletzky, R.M., and Kathariou, S. 2008. Host ranges of Listeria-specific bacteriophages from the turkey processing plant environment in the United States. Applied and Environmental Microbiology. 74: 6623–6630. https://doi. org/10.1128/AEM.01282-08
Kuenne, C., Billion, A., Mraheil, M.A., Strittmatter, A., Daniel, R., Goesmann, A., Barbuddhe, S., Hain, T., and Chakraborty, T. 2013. Reassessment of the Listeria monocytogenes pan-genome reveals dynamic integration hotspots and mobile genetic elements as major components of the accessory genome. BMC Genomics. 14: 47. https://doi.org/10.1186/1471-2164-14-47
Loessner, M.J., and Busse, M. 1990. Bacteriophage typing of Listeria species.
Applied and Environmental Microbiology. 56: 1912–1918.
Loessner, M.J., Goeppl, S., and Busse, M. 1991. Comparative inducibility of bacteriophage in naturally lysogenic and lysogenized strains of Listeria spp. by UV light and Mitomycin C. Letters in Applied Microbiology. 12: 196–199. https://doi.org/10.1111/j.1472-765X.1991.tb00538.x
López, E., Domenech, A., Ferrándiz, M.-J., Frias, M.J., Ardanuy, C., Ramirez, M., García, E., Liñares, J., and de la Campa, A.G. 2014. Induction of prophages by fluoroquinolones in Streptococcus pneumoniae: implications for emergence of resistance in genetically-related clones. PloS one. 9: e94358. https://doi. org/10.1371/journal.pone.0094358
Mammina, C., Parisi, A., Guaita, A., Aleo, A., Bonura, C., Nastasi, A., and Pontello, M. 2013. Enhanced surveillance of invasive listeriosis in the Lombardy region, Italy, in the years 2006-2010 reveals major clones and an increase in serotype 1/2a. BMC Infectious Diseases. 13: 152. https://doi. org/10.1186/1471-2334-13-152
Marini, E., Magi, G., Vincenzi, C., Manso, E., and Facinelli, B. 2016. Ongoing outbreak of invasive listeriosis due to serotype 1/2a Listeria monocytogenes, Ancona province, Italy, January 2015 to February 2016. Eurosurveillance. 21: 30217. https://doi.org/10.2807/1560-7917.ES.2016.21.17.30217
Nelson, K.E., Fouts, D.E., Mongodin, E.F., Ravel, J., DeBoy, R.T., Kolonay, J.F., Rasko, D.A., Angiuoli, S.V, Gill, S.R., Paulsen, I.T., et al. 2004. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Research. 32: 2386–2395. https://doi.org/10. 1093/nar/gkh562
Orsi, R.H., Borowsky, M.L., Lauer, P., Young, S.K., Nusbaum, C., Galagan, J.E., Birren, B.W., Ivy, R.A., Sun, Q., Graves, L.M., et al. 2008. Short-term genome evolution of Listeria monocytogenes in a non-controlled environment. BMC Genomics. 9: 539. https://doi.org/10.1186/1471-2164-9-539
Palumbo, S.A., and Williams, A.C. 1991. Resistance of Listeria monocytogenes to freezing in foods. Food Microbiology. 8: 63–68. https://doi.org/10.1016/0740- 0020(91)90017-V
Poblet-Icart, M., Bordons, A., and Lonvaud-Funel, A. 1998. Lysogeny of Oenococcus oeni (syn. Leuconostoc oenos) and study of their induced bacteriophages. Current Microbiology. 36: 365–369. https://doi.org/10.1007/s002849900324 Sekulovic, O., Garneau, J.R., Néron, A., and Fortier, L.C. 2014. Characterization of temperate phages infecting Clostridium difficile isolates of human and animal origins. Applied and Environmental Microbiology. 80: 2555–2563. https://doi.org/10.1128/AEM.00237-14
Swaminathan, B., and Gerner-Smidt, P. 2007. The epidemiology of human listeriosis. Microbes and Infection. 9: 1236–1243. https://doi.org/10.1016/j. micinf.2007.05.011
Tolvanen, R., Hellström, S., Elsser, D., Morgenstern, H., Björkroth, J., and Korkeala, H. 2008. Survival of Listeria monocytogenes strains in a dry sausage model. Journal of Food Protection. 71: 1550–1555. https://doi.org/10.4315/ 0362-028X-71.8.1550
Verghese, B., Lok, M., Wen, J., Alessandria, V., Chen, Y., Kathariou, S., and Knabel, S. 2011. comK prophage junction fragments as markers for Listeria monocytogenes genotypes unique to individual meat and poultry processing plants and a model for rapid niche-specific adaptation, biofilm formation, and persistence. Applied and Environmental Microbiology. 77: 3279–3292. https://doi.org/10.1128/AEM.00546-11
Vongkamjan, K., Benjakul, S., Vu, H.T.K., and Vuddhakul, V. 2017. Longitudinal monitoring of Listeria monocytogenes and Listeria phages in seafood processing environments in Thailand. Food Microbiology. 66: 11–19. https:// doi.org/10.1016/j.fm.2017.03.014
Vongkamjan, K., Fuangpaiboon, J., Jirachotrapee, S., and Turner, M.P. 2015. Occurrence and diversity of Listeria spp. in seafood processing plant environments. Food Control. 50: 265–272. https://doi.org/10.3153/JFHS16004 Vongkamjan, K., Fuangpaiboon, J., Turner, M.P., and Vuddhakul, V. 2016. Various ready-to-eat products from retail stores linked to occurrence of diverse Listeria monocytogenes and Listeria spp. isolates. Journal of Food Protection.
79: 239–245. https://doi.org/10.4315/0362-028X.JFP-15-361
Vongkamjan, K., Moreno Switt, A., den Bakker, H.C., Fortes, E.D., and Wiedmann, M. 2012. Silage collected from dairy farms harbors an abundance of listeriaphages with considerable host range and genome size diversity. Applied and Environmental Microbiology. 78: 8666–8675. https://doi.org/ 10.1128/AEM.01859-12
Wendlinger, G., Loessner, M.J., and Scherer, S. 1996. Bacteriophage receptors on Listeria monocytogenes cells are the N-acetylglucosamine and rhamnose substituents of teichoic acids or the peptidoglycan itself. Microbiology. 142: 985–992. https://doi.org/10.1099/00221287-142-4-985
Wright, E.E., Elliman, J.R., and Owens, L. 2013. Induction and characterization of lysogenic bacteriophages from Streptococcus iniae. Journal of Applied Microbiology. 114: 1616–1624. https://doi.org/10.1111/jam.12192
Hue Thi Kim Vu1,2, Soottawat Benjakul1, Varaporn Vuddhakul3, and Kitiya Vongkamjan1*
1Department of Food Technology, Faculty of Agro-Industry, Prince of Songkla University, Hat Yai 90110, Thailand
2Department of Veterinary Hygiene and Food Safety, National Institute of Veterinary Research (NIVR), Dong Da, Hanoi, Vietnam
3Department of Microbiology, Faculty of Science, Prince of Songkla University, Hat Yai 90110, Thailand
*Corresponding author. E-mail: kitiya.v@psu.ac.th
Total Article Views
Article history:
Received: July 16, 2018;
Revised: October 24, 2018;
Accepted: November 1, 2018

