
Identifying biofilm-forming strains of Staphylococcus epidermidis isolated from intravascular-catheterized patients by icaA and icaD genes
Gamal A. El-Sawaf, Sara M.A.I. Adlan, Dalia Metwally, Magda M. Abo-Ollo, Medhat M.A. Hamed, and Mohamed S. Abdel-Latif*Published Date : 2022-03-31
DOI : https://doi.org/10.12982/CMUJNS.2022.024
Journal Issues : Number 2, April-June 2022
Abstract Staphylococcus epidermidis has gained a substantial importance in recent years because it is one of the main causative agents of nosocomial infections. It requires a predisposed host in order to switch from a normal inhabitant of the human skin to a pathogenic flora. This study aimed to use icaA/icaD genes as biomarkers in differentiating biofilm-forming S. epidermidis, obtained from patients with intravascular catheter (IVC) infections, from other saprophytic strains. Twenty isolates of S. epidermidis obtained from 100 cases of intravascular catheter infections were investigated for the presence of the intracellular adhesion icaA and icaD genes by polymerase chain reaction (PCR) and for phenotypic biofilm production by qualitative Congo red agar assay (CRA). Results: Nine (~45%) S. epidermidis isolates out of 20 isolates collected from IVC infections were positive for both CRA (produce black colonies) and icaA/icaD genes; while 11 (~55%) S. epidermidis isolates were negative for CRA and icaA/icaD genes. Detection of icaA/icaD gene is a reliable, efficient, and more rapid tool for characterizing biofilm-forming strains of S. epidermidis.
Keywords: Staphylococcus epidermidis; icaA; icaD; biofilm
Funding: This research did not receive any specific grant from funding agencies in the public or commercial sectors.
Citation: El-Sawaf, G.A., Adlan, S.M.A.I., Metwally, D., Abo-Ollo, M.M., Hamed, M.M.A., and Abdel-Latif, M.S. 2022. Identifying biofilm-forming strains of Staphylococcus epidermidis isolated from intravascular-catheterized patients by icaA and icaD genes. CMU J. Nat. Sci. 21(2): e2022024.
INTRODUCTION
Bacterial infection has been considered as one of the main cause of implant failure. Moreover, research has been conducted to understand the mechanism by which the normal saprophytic bacteria are capable to succeed in colonizing prostheses, and cause host pathogenesis [Ziebuhr et al., 1997]. The role played by slime in implant infection is well documented when examining its prevalence in strains isolated from the normal epithelial microflora and in implant-associated infection. Accordingly, all implanted devices are susceptible to the risk of infection that associated with an increase in morbidity and mortality. Therefore, the understanding of mechanisms by which pathogens adhere and colonize a device is much greater than just a scientific interest. For instance, it is also the base for the research and development of infection-resistant materials. Therefore, in vitro studies of the pathogenesis of biomaterial-related infections are crucial [Arciola et al., 1994, Arciola et al., 1998]. Biofilms are structured communities of bacterial cells enclosed in a self-produced external matrix of polysaccharide and adherent to an inert or living surface. Establishment of a biofilm is the initial step for developing various chronic infections (such as biomaterial-associated wounds), and pulmonary infections [Fitzpatrick et al., 2005]. The complexities of the system that lead to staphylococcal biofilm formation are just beginning to be fully realized. Biofilm develops in a stepwise manner that includes cell attachment, accumulation, maturation, and detachment [Rohde et al., 2006]. Previous studies have established that biofilm formation is the most important pathogenic factor during Staph. epidermidis infection [Maraj et al., 2004, El Helou et al., 2009]. Therefore, the biofilm phenotype was used as a marker to differentiate pathogenic strains from normal strains [Caputo et al., 1987]. Several specific genes have been evaluated as potential genetic markers for invasiveness of biofilm-forming Staph. epidermidis. A special attention has been given to the ica gene locus [Roos, 2009]. It became well known that slime synthesis is controlled by the ica operon [Gerke et al., 1998]. The activation of the ica operon provoke the synthesis of the polysaccharide intracellular adhesion (PIA), a main slime component consisting of linear β-1, 6-linked glucosaminylglycans. PIA is synthesized in vitro from UDP-N-acetyl glucosamine by the enzyme N-acetyl glucosaminyl transferase, which is encoded by the intracellular adhesion (ica) locus and in particular by the icaA gene. Expression of icaA alone induces a low enzymatic activity, but coexpression of icaA with icaD leads to a significant increase in the enzymatic activity and is related to the full phenotypic expression of the capsular polysaccharide. However, molecular methods provide a direct evidence of the genetic basis of slime production; and consider complementary to the Congo red agar assay (CRA). In addition, molecular techniques for the identification of the ica genes that encode for the slime synthesis represent a very reliable tool for an accurate identification of the virulent slime-forming strains [Arciola et al., 2001a, 2001b].
The current study aimed to use icaA and icaD genes as biomarkers in differentiating biofilm-forming Staph. epidermidis, obtained from patients with intravascular catheter (IVC) infections, from other saprophytic strains.
MATERIAL AND METHODS
Bacterial strains
A total of 20 Staph. epidermidis strains were isolated from 100 infected intravascular catheters (IVC) removed from patients admitted to the Medical Research Institute hospital, Alexandria University, Egypt. Briefly, the catheters were removed, aseptically, from patients and were transported in refrigerated form to the bacteriology laboratory for bacterial infection test, shortly after arrival.
As negative control for the study; 25 Staph. epidermidis isolates were obtained from the hands of healthy volunteers, who did not attend the hospital.
Informed consents were obtained from all recruited individuals. Also, ethical approval for the study was obtained from the local ethical committee of the Medical Research Institute, University of Alexandria, Egypt.
Characterization of bacterial strains obtained from infected IVC
The distal ends of the catheters were inoculated in 1 ml of lysogeny broth (LB) and then 100 µl of the broth were inoculated onto blood agar and incubated at 37ºC under aerobic condition for 24 hr. Cultures were considered significant when the bacterial count was 103 CFU/ml.
Then, identification of Staphylococci was achieved by colonial morphology, Gram staining (spherical gram-positive cocci arranged in irregular grapelike clusters), positive catalase test, negative coagulase (tube and slide coagulase), and negative modified oxidase test. Species identification was performed using the API20E Staph (API Bio Mériux, le Balme Les Grottes, France) according to the manufacturer procedures.
Identification of gram positive Staphylococci, briefly, as following:
A. Catalase test
A colony of the culture was picked with a sterile toothpick and transferred to a glass slide. One to two drops of 3% hydrogen peroxidase were added to the colony on the glass slide. Production of gas bubbles indicated a positive reaction that is characterized for Staphylococci [McLeod and Gordon, 1923].
B. Modified oxidase test
A 1% (w/v) solution of tetra-methyl-p-phenylene-diamine dihydrochloride in certified grade dimethyl sulphoxide was used to differentiate micrococci from Staphylococci [Tarrand and Grosch, 1982]. The differentiation is based on the detection of oxidase enzyme. For a detection of oxidase enzyme a filter paper circular discs impregnated with tetra-methyl-p-phenylene-diamine dihydrochloride (oxidase reagent) in dimethyl sulfoxide (DMSO) are used. DMSO aids in the permeability of cells to the oxidase reagent. In presence of atmospheric oxygen, the oxidase enzyme reacts with the oxidase reagent and cytochrome C to form the colored compound, indophenol indicated as blue or purple blue coloration on the disc after the introduction of bacterial colony on the disc. The development of blue or purple blue on the disc within 2 min is considered as a positive result, and that is characterizing the micrococci. Staphylococci should yield a negative result (i.e. no color change) [Tarrand and Grosch, 1982].
C. Clumping and free coagulase test, and mannitol fermentation
Catalase positive and oxidase negative Staphylococci were, first, tested for; i) clumping factor production by slide coagulase test; briefly, a staphylococcal colony was emulsified in a drop of water on a microscope slide to form a smooth milky suspension, then a straight loop was dipped into the undiluted citrated human plasma and withdrawn. The adhering traces of plasma were stirred into the Staphylococcal suspension. A visible coarse clumping within 10 seconds was read as positive result that characterizes Staph. aureus; while, the negative results characterize Staph. epidermidis and staph. haemolyticus [Dickson and Marples, 1986]. ii) Free coagulase production by tube coagulase test; briefly, 1 ml of 1:6 dilution of human plasma was placed in small tube, then, a colony of staphylococci under test was emulsified in the tube. Then, tube was incubated at 37°C for up to 4 hours. Tube was examined for clot formation by tilting. Negative tubes were left 30 min at room temperature and re-examined for delayed clot formation [Dickson and Marples, 1986].
Second, catalase positive and oxidase negative Staphylococci were inoculated on mannitol salt agar (MSA) containg mannitol 1%, NaCl 7.5% with phenol red as indicator of acid production. Mannitol salt agar (MSA) has been used for many years as selective differential media for the isolation of S. aureus on the high salt content of medium. Thus, the differentiation between S. aureus and the coagulase negative Staphylococci (CONS) will be according to fermentation of mannitol, where CONS are mannitol negative (non-fermenter) [Shittu et al., 2006].
Preparation of Bacterial Stock
For bacterial storage and revival; briefly, 1 ml of fresh saturated bacterial culture grown on lysogeny broth (LB) was added to 1 ml of sterile glycerol solution in screw-capped cryo-tube. The tubes were stored at -20°C. For bacterial revival, one loopful was streaked over blood agar and incubated at 37°C [Howard, 1956].
Identification of Staph. epidermidis by Analytical Profile Index (API 20E Test)
Isolated staphylococci were sub-cultured on blood agar and incubated for 24 hours at 37ºC. Afterward, the organism was biochemically identified using the API 20E Staph system (API Bio Mériux, le Balme Les Grottes, France) according to the manufacturer procedures.
Determination of biofilm formation by Congo Red Agar (CRA)
Staph. epidermidis isolates were cultured onto Congo red agar plates (CRA; 1 L Brain-Heart infusion agar, Oxoid, UK; 36 g Saccharose, Sigma, Germany; 0.8 g Congo red, S D Fine-Chem Limited, India). Plates were incubated for 24 hours at 37ºC. Biofilm producing strain formed reddish-black colonies with a rough dry and crystalline consistency on Congo red agar, whereas a non-biofilm producing strain developed pinkish-red smooth colonies [Freeman et al., 1989].
Molecular detection of icaA/icaD genes
Staph. epidermidis isolates were subcultured overnight at 37°C on blood agar for 24 hours. Few colonies were emulsified in 200 µl sterile distilled water to produce a heavy suspension. DNA was extracted from a bacteria isolates using the commercially available QIAamp MiniKit according to manufacturer procedures (Qiagen, Hilden, Germany). DNA was amplified by conventional PCR technique using two different sets of primers as previously described [Arciola et al., 2001a]. icaA forward primer, 5’-TCTCTTGCAGGAGCAATCAA-3’ and icaA reverse primer, 5’-TCAGGCACTAACATCCAGCA-3’; which amplify a specific 188-base pair fragment. icaD forward primer, 5’-ATGGTCAAGCCCAGACAGAG-3’, and icaD reverse primer, 5’-CGTGTTTTCAACATTTAATGCAA-3’; which amplify a specific 198-base pair fragment. Ready to go PCR beads (Amersham Pharmacia Biotech) are designed as pre mixed pre-dispensed reaction for performing PCR amplification. PCR beads are provided as dried beads that are stable at room temperature. Each bead contains all the necessary reagents, except primers and template, for performing a 25 µl PCR amplification reaction. Each individual PCR reaction (25 µl) contains: 1.5 unit of taq DNA polymerase, 10 mM Tris HCl (pH 9), 50 mM MgCl2, 200 mM of each dNTP and a stabilizer. Then, DNA and primers were added onto each PCR reaction. The contents of each reaction tube were mixed gently then placed in a Perkin Elmer 9600 thermocycler to perform the PCR amplification cycles. The amplified DNA products were run on 2% agarose containing ethidium bromide (0.5 µg/ml) by electrophoresis and visualized on UV transilluminator for positive bands; taking into consideration that a similar volumes of amplified DNA products of all samples were loaded in all lanes as well as standard DNA ladder, so that the comparison of bands becomes more feasible.
RESULTS
A total of 100 bacterial isolates from IVC infections were collected from patients admitted to the hospital of Medical Research Institute, Alexandria. After overnight incubation of bacterial culture, the organisms were identified and speciated according to standard microbiological techniques. Gram-positive cocci, catalase positive, and oxidase negative Cocci colonies were further examined for coagulase test and sub-cultured on mannitol salt agar (MSA). The identified 20 S. epidermidis isolates (coagulase negative and mannitol non-fermenter) were examined for API 20E Test, Congo red agar method and molecular detection of icaA/icaD genes using PCR technique.
Bacteria isolates from 100 IVC infections
Table 1 shows that, out of the 100 cases of IVC infections 40 (40%) cases were Gram-negative, 60 (60%) cases were Gram-positive. Twenty (33.33%) out of the 60 Gram-positive isolates were Staphylococcus epidermidis, 5 (8.33%) were S. haemolyticus, and 10 (16.66%) were S.aureus, while 25 (41.66%) were micrococci.
Table 1. Bacteria isolates from 100 IVC infected patients.
|
Bacteria |
Number |
% |
|
S. epidermidis |
20 |
20 |
|
S. haemolytics |
5 |
5 |
|
S. aureus |
10 |
10 |
|
Micrococci |
25 |
25 |
|
Gram negative |
40 |
40 |
|
Total |
100 |
100 |
Coagulase production
Table 2 shows that, among the 35 Staphylococci isolates, 10 (28.57%) S. aureus isolates were positive for both free coagulase and clumping factor while, 25 (71.42%) S. epidermidis and S. haemolyticus were negative for free and clumping coagulase test (as detected by slide and tube coagulase test).
Table 2. Coagulase production in the 35 Staphylococci isolates.
|
Coagulase Production |
Free Coagulase |
Clumping Factor |
||
|
Number |
% |
Number |
% |
|
|
Coagulase Positive |
10 |
28.57 |
10 |
28.57 |
|
(Staph. aureus) |
|
|
|
|
|
Coagulase Negative Staphylococci (CONS) |
25 |
71.42 |
25 |
71.42 |
|
Total |
35 |
100 |
35 |
100 |
Mannitol fermentation
Table 3 shows that out of 35 Staphylococci isolate only 10 (28.57%) S. aureus isolates were mannitol fermenter while 25 (71.42%) were isolated as mannitol non fermenter (S. epidermidis and S. haemolyticus).
Table 3. Mannitol fermentation in the 35 Staphylococci isolates.
|
Mannitol Fermentation |
Number |
% |
|
Staph. aureus (Fermentor) |
10 |
28.57 |
|
Coagulase Negative Staphylococci (CONS) (Non-Fermentor) |
25 |
71.42 |
|
Total |
35 |
100 |
Staph. epidermidis Identification
Table 4 shows that, 20 (80%) isolates out of 25 coagulase and mannitol negative Staphylococci that examined by Analytical Profile Index (API 20E Test), were identified as Staph. epidermidis; and 5 (20%) isolates were identified as Staph. haemolyticus.
Table 4. Staph. epidermidis Identification by API 20E Test.
|
API Staph. Identification |
Number |
% |
|
Staph. epidermidis |
20 |
80 |
|
Staph. haemolyticus |
5 |
20 |
Detection of Biofilm formation by Congo Red Agar (CRA)
Phynotypic production of biofilm, for the 20 S. epidermidis isolates, was assessed by Congo Red Agar (CRA) culture in which the black colonies are considered as biofilm producers and red colonies are considered as biofilm non-producers.
Table 5 shows that, 9 (45%) isolates out of 20 S. epidermidis isolates were biofilm producers (with black colonies on CRA); while the other 11 (55%) isolates were biofilm non-producers (with red colonies on CRA).
On the contrary, the 25 S. epedermidis isolates from healthy volunteers showed that, 3 (12%) isolates were biofilm producers and the other 22 (88%) isolates were biofilm non-producers (Table 5).
Table 5. Detection of Biofilm formation by Congo Red Agar (CRA).
|
Biofilm Formation (CRA culture) |
IVC isolated (20 patients) |
Health Volunteers’ isolated (25 control individual) |
||
|
Number |
% |
Number |
% |
|
|
Biofilm Producers |
9 |
45 |
3 |
12 |
|
Biofilm Non-Producers |
11 |
55 |
22 |
88 |
|
Total |
20 |
100 |
25 |
100 |
Detection of icaA/icaD genes
The presence of icaA/icaD genes was detected by PCR; which is considered as the gold standard method for detection of S. epidermidis isolates that produce biofilms. According to the PCR results, 9 (45%) isolates out of 20 S. epidermidis isolates were positive for icaA/icaD genes (figure 1), that means they are biofilm producers; while the other 11 (55%) isolates were negative for icaA/icaD genes, that means they are biofilm non-producers. On the contrary, the 25 S. epedermidis isolates from healthy volunteers showed that only 1 (4%) isolate was positive for icaA/icaD genes and thus considered as biofilm producer; while the other 24 (96%) isolates were negative for icaA/icaD genes and thus considered as biofilm non-producers (data not shown).
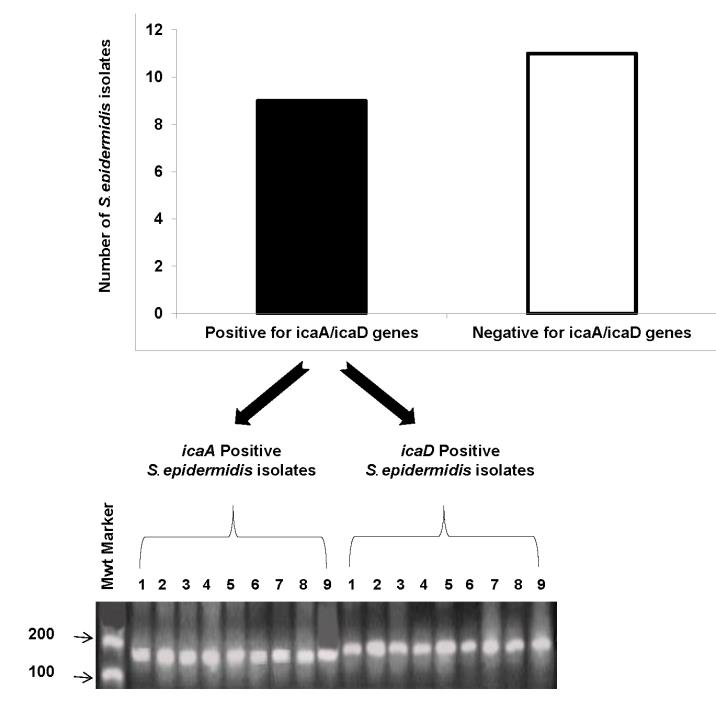
Figure 1. A) shows nine S. epidermidis isolates which are positive for both icaA and icaD genes, and eleven isolates are negative for both icaA and icaD genes.
B) shows bands of icaA and icaD genes of S. epidermidis isolates on agarose gel. icaA gene is amplified at 188 bp, and icaD at 198 bp. Numbers from 1 – 9 reflect positive isolates for icaA gene; and from 1’ – 9’ show that same isolates are also positive for icaD gene.
Biofilm formation by CRA culture versus icaA/icaD gene detection
A comparison has been made between CRA culture for biofilm forming S. epidermidis isolates, and PCR for icaA/icaD gene detection in the same isolate. And, that was run on both IVC patients with S. epidermidis infection, and S. epidermidis isolated from healthy volunteers (as control). It was noticed that, there is a concordance between the results obtained by CRA culture for S. epidermidis with biofilm formation properties, and the results obtained by PCR for detection of icaA/icaD genes that characterize the biofilm formation properties in IVC patients with S. epidermidis infection (Figure 2). S. epidermidis isolated from healthy volunteers showed a little variation between CRA and icaA/icaD gene detection in 3 isolates with biofilm formation properties (Figure 2).
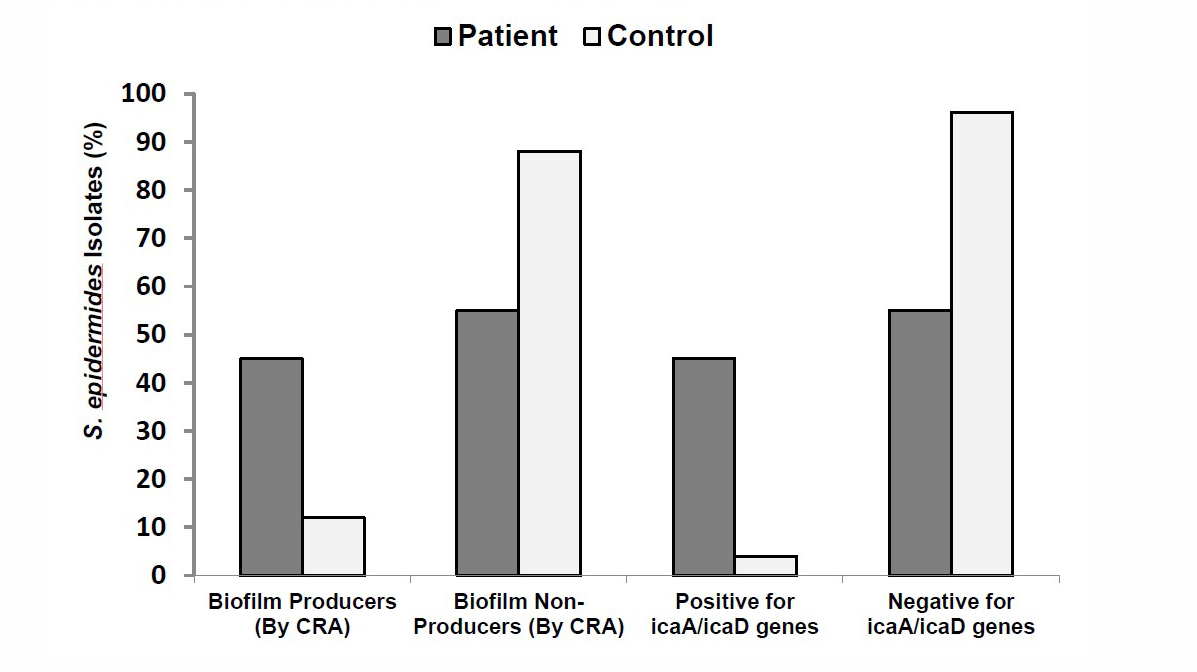
Figure 2. Biofilm formation by CRA culture versus icaA/icaD genes detection.
DISCUSSION
In the current study, the presence of icaA/icaD genes, responsible for biofilm formation, was investigated in 20 Staph. epidermidis isolates from IVC infected patients by a simple, rapid and highly specific PCR based procedure. The same strains were also analyzed by an optimization of CRA test. Due to the difficulty of chromatic evaluation involved in the original method, a print six-color reference scale was adopted to support the phase of colony identification [Arciola et al., 2002]. Finally, the color classification of each strain performed by the CRA test was compared with the more reliable information derived from the molecular analysis of icaA/icaD genes.
Biofilms are adherent layers of bacteria, characterized by cells that are permanently attached to surfaces. Bacteria communicate with each other within the biofilm by a quorum sensing. Biofilms produce a matrix of extracellular polymeric substances (EPS) called polysaccharide intercellular adhesin (PIA) [Donlan and Costerton, 2002]. PIA is synthesized from UDP-N-acetylglucosamine by N-acetylglucosaminyltransferase which is encoded by ica gene [Gerke et al., 1998]. The products of ica gene (intercellular adhesion) mediates the synthesis of PIA, which are organized in an operon structure. This operon contains the icaADBC genes, in addition to the icaR gene, which has a regulatory function and is transcribed in the opposite direction. The icaADBC locus is widespread in Staph. epidermidis isolates [Heilmann et al., 1996]. Expression of ica genes promotes a significant increase in N-acetylglucosaminyltransferase, which results in biosynthesis of PIA [Gerke et al., 1998].
Results revealed that 9 (45%) isolates out of the 20 Staph. epidermidis isolates were tested positive for icaA/icaD genes; and 11 (55%) were tested negative for icaA/icaD genes. Same results obtained by CRA test; where, 9 (45%) isolates out of 20 Staph. epidermidis isolates were biofilm producers (with black colonies in CRA); while the other 11 (55%) isolates (same isolates that were tested negative for icaA/icaD genes) were biofilm non-producers (with red colonies in CRA). On the contrary, the 25 Staph. epidermidis isolates collected from healthy volunteers (as control) were tested for icaA/icaD genes by PCR technique; and showed that 1 (4%) isolate was found to be positive for icaA/icaD genes, while the other 24 (96%) isolates were negative. However, when those 25 Staph. epidermidis control isolates were tested by CRA they showed 3 (12%) isolates were biofilm producers and the other 22 (88%) isolates were biofilm non-producers. Such variation could be explained as; the analysis of ica genes can confirm that, transition from positive slime forming isolates to negative occurs when colonies on CRA plates turn from a black to a pink/red color. The chromatic scale was found to be a feasible tool for better discrimination between these two extreme tones [Arciola et al., 2002]. Each single colony formed on the agar could be monitored for the development of possible variants. In a number of bacterial strains, at 48 hours of incubation, the formation of pink/red colored spikes within black colonies could be evidenced by this technique [Arciola et al., 2002]. Concerning the nature of such spikes, there is some experimental proof which supports the development of new clones referred to as biofilm-negative, which have lost both icaA/icaD genes [Arciola et al., 1994, Ziebuhr et al., 1997]. Moreover, similar changes encountered in bacteria derived from a single producer strain were attributed to phenotypic and not to genotypic variations [Arciola et al., 1994, Ziebuhr et al., 1997].
Aricola et al., [2001a, 2001b, 2002] indicated a fine consistency between PCR method for icaA/icaD gene detection and the CRA method for slime formation. Their reported data indicate the important role of icaA/icaD genes as a virulence marker in Staphylococcal infections from intravenous catheters. In addition, their results showed that CRA method has a good reliability, especially when supported by a chromatic scale. Regardless the variations in sample size (ranging from 15 to 113 Staph. epidermidis strains) that noticed in previously published studies, there was a consensus that the results obtained by PCR method do agree with the results obtained by CRA method, with considerable margin of reproducibility and performance [Aricola et al., 2001a, 2001b, 2002].
On the contrary, Chaieb et al., [2005] investigated the presence of the intracellular adhesion genes (icaA/icaD) by PCR, and phenotypic biofilm production by qualitative CRA assay. A total of 32 Staph. epidermidis strains were identified from dialysates and needles 4 hour after the initiation of dialysis. Qualitative biofilm production revealed that 16 (50%) strains were biofilm positive on CRA plates. Twenty-three isolates were PCR positive for the icaA/icaD genes; out of them 15 isolates were biofilm positive on CRA and 8 were biofilm negative. Only one isolate, from the 32 Staph. epidermidis strains, was icaA/icaD negative but still forming slime. Accordingly, they reported that the ability of Staph. epidermidid to produce slime is not associated with the presence of icaA/icaD genes. One interpretation could be raised for that finding which is the absence of chromatic scale for verifying those transformed clones from slime forming to non- forming ones (i.e. from black to pink/red colony).
CONCLUSION
In clinical settings, the identification of bacteria using the classical method and determination of their biofilm formation capability, generally, require few days of time, whereas, the use of PCR assay for the detection of icaA/icaD genes is more rapid and efficient. Therefore, the molecular epidemiological tools are useful for understanding the transmission patterns to control nosocomial infection. Moreover, that can help clinicians to determine the species involved in suspected Staphylococcal sepsis to evaluate biofilm formation, which is crucial for deciding treatment.
ACKNOWLEDGEMENTS
Authors appreciate the grate cooperation offered at the department of Diagnostic and Molecular Microbiology, Medical Research Institute, University of Alexandria, Egypt, for their help and support. Also, authors thank all members of Medical Laboratory Technology Department, Faculty of allied Medical Science, Pharos University in Alexandria, Egypt.
AUTHOR CONTRIBUTIONS
Gamal A. El-Sawaf, Sara M.A.I. Adlan, Dalia Metwally, Magda M. Abo-Ollo, Medhat M. A. Hamed, and Mohamed S. Abdel-Latif assisted in conducting the experiments, performed the statistical analysis and data visualization and wrote the manuscript. All authors have read and approved of the final manuscript.
CONFLICT OF INTEREST
The authors declare that they hold no competing interests.
REFERENCES
Arciola, C.R., Caramazza, R., and Pizzoferrato, A. 1994. Related articles in vitro adhesion of Staphylococcus epidermidis on heparin-surface modified intraocular lenses. Journal of Cataract & Refractive Surgery. 20: 158-161.
Arciola, C.R., Montanaro, L., Caramazza, R., Sassoli, V., and Cavedagna, D. 1998. Inhibition of bacterial adherence to a high-water-content polymer by a water-soluble, nonsteroidal, anti-inflammatory drug. Journal of Biomedical Materials Research. 42: 1-5.
Arciola, C.R., Collamati, S., Donati, E., and Montanaro, L. 2001a. A rapid PCR method for the detection of slime-producing strains of Staphylococcus epidermidis and Staphylococcus aureus in periprosthesis infections. Diagnostic Molecular Pathology. 10: 130-137.
Arciola, C.R., Baldassarri, L., and Montanaro, L. 2001b. Presence of icaA and icaD genes and slime production in a collection of Staphylococcal strains from catheter-associated infections. Journal of Clinical Microbiology. 39: 2151-2156.
Arciola, C.R., Campoccia, D., Gamberini, S., Cervellati, M., Donati, E., and Montanaro, L. 2002. Detection of slime production by means of an optimized Congo red agar plate test based on a colorimetric scale in Staphylococcus epidermidis clinical isolates genotyped for ica locus. Biomaterials. 23: 4233-4239.
Caputo, G.M., Archer, G.L., Calderwood, S.B., DiNubile, M.J., and Karchmer, A.W. 1987. Native valve endocarditis due to coagulase negative Staphylococci. Clinical and microbiologic features. American Journal of Medicine. 83: 619-625.
Chaieb, K., Mahdouani, K., and Bakhouf, A. 2005. Detection of icaA and icaD loci by polymerase chain reaction and biofilm formation by Staphylococcus epidermidis isolated from dialysate and needles in a dialysis unit. Journal of Hospital Infection. 61: 225-230.
Dickson, J.I. and Marples, R.R. 1986. Coagulase production by strains of Staphylococcus aureus of differing resistance characters: a comparison of two traditional methods with a latex agglutination system detecting both clumping factor and protein A. Journal of Clinical Pathology. 39: 371-375.
Donlan, R.M. and Costerton, W. 2002. Biofilms: Survival mechanisms of clinically relevant Microorganisms. Clinical microbiological review. 5: 167-193.
El Helou, O.C., Berbari, E.F., and Osmon, D.R. 2009. Osteomyelitis and Other Bone and Joint Infections, pp 424-443. In Crossley KB, Jefferson KK, Archer GL, Fowler VG (eds.), Staphylococci in Human Disease, 2nd Ed. Blackwell Publishing Ltd.
Freeman, J., Falkiner, F.R., and Keane, C.T. 1989. New method for detecting slime production by coagulase negative Staphylococci. Journal of Clinical Pathology. 42: 872-874.
Fitzpatrick F, Humphreys H, O’Gase JP. 2005. The genetics of Staphylococcal biofilm formation. J Clin Microbiol Infect. 11:967-973.
Gerke, C., Kraft, A., Sussmuth, R., Schweitzer, O., Gotz, F., Kraft, A., et al. 1998. Characterization of the Nacetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesion. Journal of Biological Chemistry. 273: 18586-18593.
Heilmann, C., Schweitzer, O., Gerke, C., Vanittanakom, N., Mack, D., and Gotz, F. 1996. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Molecular Microbiology. 20: 1083–1091.
Howard, D.H. 1956. The preservation of bacteria by freezing in glycerol broth. Journal of Bacteriology. 71: 625.
Maraj, S., Jacobs, L.E., Maraj, R., and Kotler, M.N. 2004. Bacteremia and infective endocarditis in patients on hemodialysis. American Journal of the Medical Sciences. 327: 242-249.
McLeod, J.W. and Gordon, J. 1923. Catalase production and sensitiveness to hydrogen peroxide amongst bacteria: with a scheme for classification based on the properties. Journal of Pathology and Bacteriology. 26: 326-331.
Rohde, H., Mack, D., Christner, M., Burdelski, C., Frannke, G., and Knobloch, J.K. 2006. Pathogenesis of Staphylococcal device related infections: from basic science to new diagnostic, therapeutic and prophylactic approaches. Reviews and Research in Medical Microbiology. 17: 45-54.
Roos, K.L. 2009. Central Nervous System Infections, pp. 395-413. In Crossley KB, Jefferson KK, Archer GL, Fowler VG (eds.), Staphylococci in Human Disease, 2nd Ed. Blackwell Publishing Ltd.
Shittu, A., Lin, J., Morrison, D., and Kolawole, D. 2006. Identification and molecular characterization of mannitol salt positive, coagulase negative Staphylocci from nasal sample of medical personal and students. Journal of Medical Microbiology. 55: 317-324.
Tarrand, J.J. and Grosch, D.H. 1982. Rapid, modified oxidase test for oxidase-variable bacterial isolates. Journal of Clinical Microbiology. 16: 772-774.
Ziebuhr, W., Heilmann, C., and Götz, F. 1997. Detection of the intracellular adhesion gene cluster (ica) and phase variation in Staphylococcus epidermidis blood culture strains and mucosal isolates. Infect Immun. 65: 890-896.
OPEN access freely available online
Chiang Mai University Journal of Natural Sciences [ISSN 16851994]
Chiang Mai University, Thailand
https://cmuj.cmu.ac.th
Gamal A. El-Sawaf 1, Sara M.A.I. Adlan 1, Dalia Metwally 1, Magda M. Abo-Ollo 2, Medhat M.A. Hamed 3, and Mohamed S. Abdel-Latif 4, *
1 Department of Diagnostic and Molecular Microbiology, Medical Research Institute, University of Alexandria, Egypt.
2 Department of Anesthesia, Medical Research Institute, University of Alexandria, Egypt.
3 Department of Experimental and Clinical Surgery, Medical Research Institute, University of Alexandria, Egypt.
4 Department of Medical Laboratory Technology, Faculty of Applied Health Sciences Technology, Pharos University in Alexandria, Egypt.
Corresponding author: Mohamed S. Abdel-Latif, E-mail: mohamed.abdellatif@pua.edu.eg
Total Article Views
Editor: Veerasak Punyapornwithaya
Chiang Mai University, Thailand
Article history:
Received: November 30, 2021;
Revised: January 11, 2022;
Accepted: January 21, 2022;
Published online: January 27, 2022

