
Computational Study of Garlic Compounds as Potential Anti-Cancer Agents for the Inhibition of CCR5 and CXCR4
Balqis Balqis, Betty Lukiati, Mohamad Amin, Siti Nur Arifah, Mochammad Fitri Atho’illah, and Nashi Widodo*Published Date : 2022-01-21
DOI : https://doi.org/10.12982/CMUJNS.2022.012
Journal Issues : Number 1, January-March 2022
Abstract Current cancer treatment methods are still inadequate due to the complexity of the cancer progression mechanism, which involves multiple genes, proteins, and signaling pathways. The discovery and validation of novel anticancer compounds remains challenging. Garlic has many medicinal properties that can combat various diseases. Organosulfur present in garlic has been shown to induce apoptosis in cancer cells; however, the underlying mechanism of action of non-organosulfur compounds from garlic in controlling cancer cells remains unclear. The present study aimed to analyze the efficacy of organosulfur and non-organosulfur compounds, including the flavonoid, terpenoid, and saponin groups, as inhibitors of C-C chemokine receptor type 5 (CCR5) and C-X-C chemokine receptor type 4 (CXCR4), which play significant roles in the progression of cancer. To determine the interactions between the active compounds of garlic and these receptors (CCR5 and CXCR4), molecular docking was performed using the PyRx v.0.8 software. Amino acid residues were analyzed and visualized using Biovia Discovery Studio and PyMol, respectively. Non-organosulfur compounds exhibited better results than the organosulfur compounds in binding affinity analysis. Tigogenin (from the saponin group) is considered to be a CCR5 inhibitor, while lupeol (from the terpenoid group) is considered to be a CXCR4 inhibitor. In conclusion, our results suggest that garlic compounds could be promising inhibitors of CCR5 and CXCR4, which are highly expressed in cancer. However, further research is needed to validate the in vitro and in vivo activities of garlic compounds for the inhibition of cancer progression.
Keywords: Anticancer agents, CCR5, CXCR4, Garlic, Organosulfur compounds, Non-Organosulfur compounds]
Funding: This research did not receive any specific grant from funding agencies in the public, commercial, or not for profit sectors.
Citation: Balqis, B., Lukiati, B., Amin, M., Arifah, S.N., Atho’illah, M.F., Widodo, N. 2022. Computational study of garlic compounds as potential anti-cancer agents for the inhibition of CCR5 and CXCR4. CMU J. Nat. Sci. 21(1): e2022012.
INTRODUCTION
Cancer is the leading cause of death worldwide (Torre et al., 2016). Around 17.2 million cancer cases have been reported globally, with 8.9 million deaths in 2016, and the number of cases further increased by 1.05% in 2018 (Global Burden of Disease Cancer Collaboration, 2018; World Health Organization, 2018). Surprisingly, in the United States, the number of deaths caused by cancer has started to surpass those caused by cardiovascular diseases (Heron and Anderson, 2016). In Indonesia, breast cancer was reported as the leading cause of death among other cancer cases by 16.6%, followed by cervix uteri and lung cancer with a percentage of 9.2% and 8.8%, respectively (The Global Cancer Observatory, 2020).
However, the current cancer treatment strategies remain inadequate (Allemani and Coleman, 2017), and there is a lack of comprehensive evidence for the combination of anticancer drugs and herbal medicines (Cheng et al., 2018; Jermini et al., 2019). Interestingly, most cancer patients now choose complementary and alternative medicine (CAM) to enhance their quality of life, increase their probability of survival, and reduce the side effects of treatment (Chan et al., 2011; Smith et al., 2014; Wu et al., 2016). Drug combinations involve complicated interactions among multiple genes, proteins, and pathways at the pharmacological and pharmacokinetic levels. The drug interactions need to be studied further, as the database of drug-gene–protein interactions is still incomplete (Hu et al., 2016). Therefore, there is an urgent need to discover and validate novel anticancer compounds, which remains a challenge for scientists worldwide.
Garlic (Allium sativum) is considered to be one of three top-selling botanical products in United States retail outlets (Wachtel-Galor and Benzie, 2011) and the second most used compound in complementary medicine due to its various medicinal properties (Amagase, 2006.; Varshney and Budoff, 2016). Garlic has low toxicity (Lestari and Rifai, 2019) and possesses anti-inflammatory (Arifah et al., 2020), immunomodulatory (Lestari and Rifa’i, 2019; Lestari et al., 2020), and anticancer properties (Cao et al., 2014). However, many studies using garlic as an anticancer agent have only focused on its organosulfur compounds. Diallyl trisulfide (DATS) is considered the primary anticancer organosulfur compound in garlic (Wang et al., 2010; Shin et al., 2014). Our previous study reported that garlic also contains relatively high levels of flavonoids, terpenoids, and saponins (Balqis et al., 2018). Moreover, the association between the non-organosulfur and organosulfur compounds in garlic have not yet been studied.
Chemokines and their receptors play critical roles in the inflammatory response mechanisms and help cancer cells migrate to the site of metastasis. C-C chemokine receptor type 5 (CCR5), one of the members of the G protein-coupled receptors (GPCRs) family that binds to multiple ligands, mainly the chemokine (C-C motif) ligand 5 (CCL5), which is highly correlated with immune cell infiltration and promotes tumorigenesis (Jiao et al., 2019; Zhang et al., 2019). In contrast, C-X-C chemokine receptor type 4 (CXCR4) is highly expressed in various human cancer cells. The binding of CXCR4 with its ligand, stromal derived factor-1 (SDF-1), is known to promote the proliferation and survival of cancer cells (Wang et al., 2006; Weitzenfeld and Ben-Baruch, 2014a). To date, the CCR5-CCL5 and CXCR4-SDF-1 pathways have been highlighted due to their tumor-promoting effects. Indeed, the inhibition of these two pathways may aid in the identification of novel anticancer candidates in the future (Weitzenfeld and Ben-Baruch, 2014a; Kalpana et al., 2019). The current study investigated the potential of organosulfur and non-organosulfur compounds to act as inhibitors of CCR5 and CXCR4. We aimed to elucidate the molecular docking mechanisms of organosulfur and non-organosulfur compounds as inhibitor of cancer cell proliferation. We hypothesized that these two major compounds in garlic may synergistically inhibit CCR5 and CXCR4, which may in turn improve the efficacy of cancer treatment.
MATERIALS AND METHODS
Preparation of proteins and ligands
The protein for docking analysis was obtained from the Protein Data Bank (PDB) (https://www.rcsb.org/) with PDB ID 4MBS for CCR5 and PDB ID 3OE8 for CXCR4. CCR5 protein (4MBS) was used as a protein ligand according to a preliminary experiment conducted by Lau et al. (2015), who used the same protein of CCR5 (4MBS) and performed the molecular docking analysis of chemokine receptor CCR5 with ligand dimethyl-[[4-[[3-(4-methylphenyl) -8, 9-dihydro-7H-benzo[7] annulene-6-carbonyl]amino]phenyl]methyl]-(oxan-4-yl)azanium (TAK-779). In the other hand, the selected CXCR4 was used based on a previous study by García-Cuesta et al. (2019) on the interactive role of increased expression of CXCR4 in tumor progression (Bohn et al., 2009; García-Cuesta et al., 2019). Water and ligands inside the protein were removed using the PyMoL software version 1.7.4.5 Edu (Schrӧdinger Inc., LLC). Structure of the ligands in the sdf format used for analysis was obtained from PubChem (https://pubchem.ncbi.nlm.nih.gov/) (Table 1).
The three-dimensional structures of organosulfur compounds, flavonoids, and terpenoids were converted into .pdb using PyMOL, while saponins were converted using the Open Babel GUI software version 2.4.1 (O’Boyle et al., 2011). The saponin group was converted using the Open Babel GUI software as the structures needed to be converted into a 3-Dimensional (3D) format first. Drug inhibitor for CCR5 used (TAK-779) (CID 183790) (Lau et al., 2015), while IT1t (CID 25178351) was used as a CXCR4 inhibitor control drug (Debnath et al., 2013).
Table 1. List of ligand and protein for molecular docking.
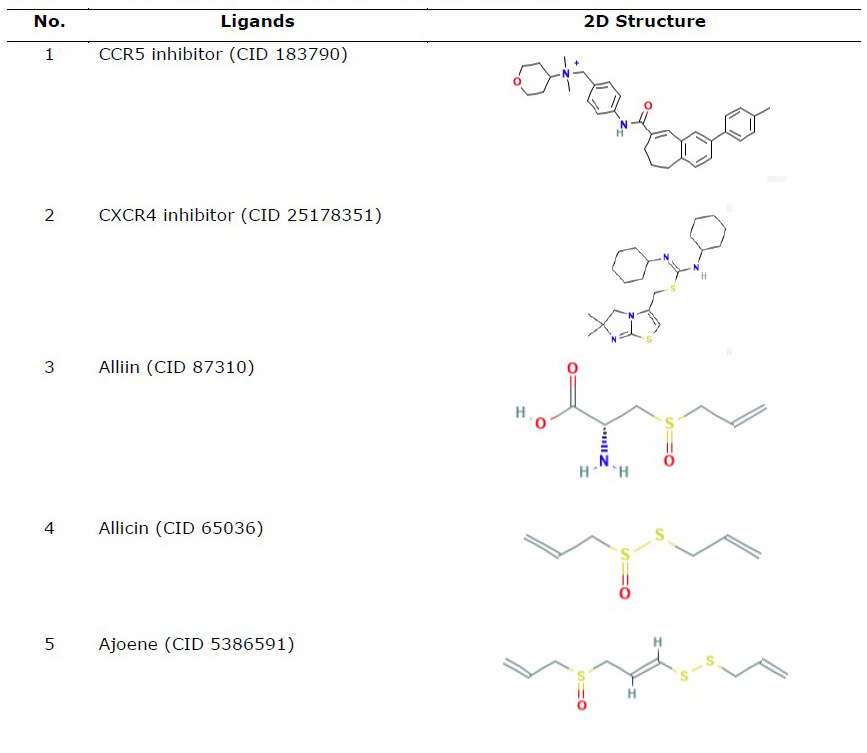
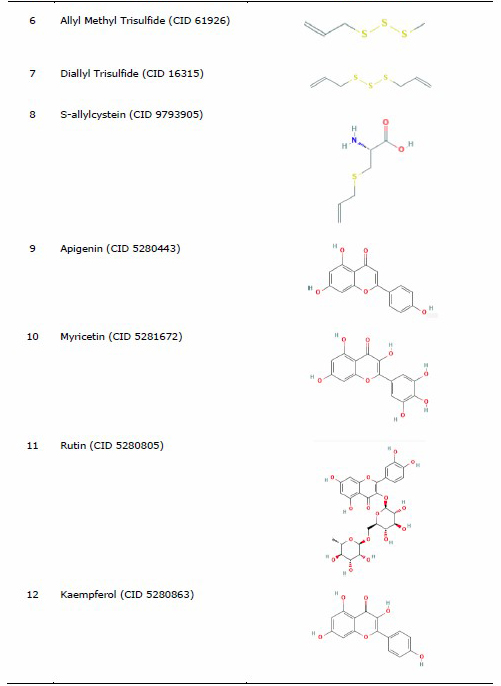
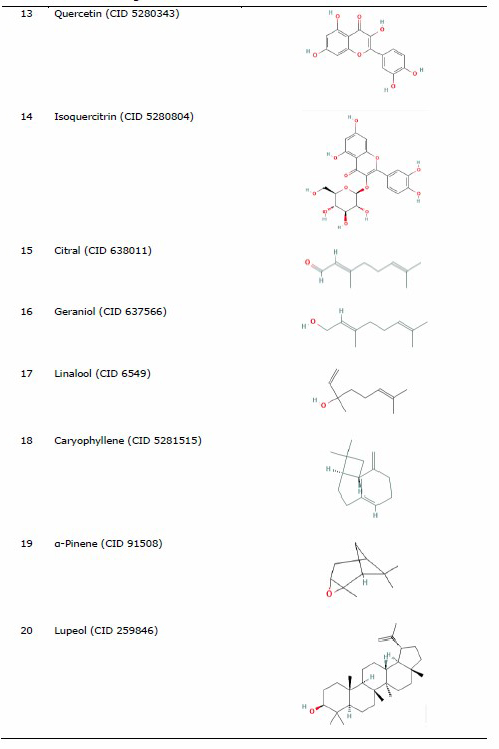
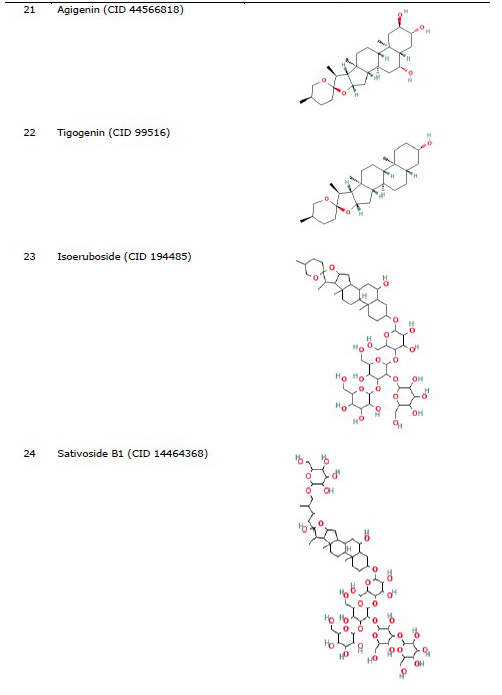
Molecular docking of ligand-protein
The interactions between the ligands and proteins were analyzed using Autodock Vina with PyRx v.0.8 (https://pyrx.sourceforge.io) (Dallakyan and Olson, 2015; Trott and Olson, 2010). Before docking, the Open Babel GUI was used to minimize the ligands with the aim of minimizing the energy of these compounds; thus, the protein-ligand interaction distance results are accurate to the level of quantum chemical calculations (Mirzaei et al., 2015). Docking was performed by binding the ligand to the active site of the protein. The center of active site for each protein was obtained by reverse docking using drug inhibitor first, TAK-779 for CCR5, and IT1t for CXCR4. Reverse docking using drug inhibitor as ligand with the aim to discover alternate drug candidate for protein which involved in specific pathway of disease. Reverse docking is utilized to identify the potential targets from the vast number of proteins/receptors by examining their known ligands or crystal structures (Huang et al., 2018; Pertami et al., 2021). Reverse docking initiated with docking between protein and drug inhibitor using blind docking. After the drug inhibitor attached to the active site of protein, then continuing to be docking with active compounds from garlic. Reverse docking using each drug inhibitor resulted the center of CCR5 active site was X: 157.9064, Y: 106.5949, Z: 19.4361, with dimensions at X: 24.8522, Y: 13.0475, Z: 15.0708 Å, while the center of CXCR4 was at X: 48.6176, Y: -0.1687, Z: 17.2453, with the dimensions of active site as X: 26.3700, Y: 20.8968, and Z: 27.9578 Å. Binding affinity values in kcal/mol were analyzed as a result of the bonding strength between the ligand and the protein. Visualization of docking and bonding interactions was performed using BIOVIA Discovery Studio (Dassault Systèmes BIOVIA, 2015).
Protein interactions and networks
Analysis of protein interactions with CCR5 and CXCR4 was performed using the Biological General Repository for Interaction Datasets (BioGRID) database (https://thebiogrid.org/). Protein networks were analyzed using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (http://string-db.org/) (Szklarczyk et al., 2015). Protein interactions by BioGRID help to determine the proteins that interacted with CCR5 which involved in cancer progression via various mechanisms or pathways. Meanwhile, STRING was used to determine the proteins (decided by researchers) that were involved in specific pathways, such as the cancer pathway, together with the list of genes in those pathways.
RESULTS
Molecular docking analysis
The binding affinity value is indicated based on the Gibbs free energy (ΔG) between the protein ligands. ΔG is negative when the system reaches equilibrium at a constant temperature and pressure (Du et al., 2016). The bonding strength of the protein-ligand is determined by the magnitude of the negative ΔG; thus, it could be considered to measure the stability of any given protein-ligand complex or the binding affinity of a ligand to a given acceptor alternatively (Gilson and Zhou, 2007)
Table 2. Binding affinity between active compounds from Garlic over CCR5 and CXCR4.
|
Ligand Name |
Binding Affinity (kcal/mol) |
|
|
CCR5 |
CXCR4 |
|
|
CCR5 inhibitor |
-10.8 |
- |
|
CXCR4 inhibitor |
- |
-8.6 |
|
Organosulfur |
|
|
|
Alliin |
-4.9 |
-5.2 |
|
Allicin |
-4.0 |
-4.1 |
|
Ajoene |
-4.6 |
-4.7 |
|
Allyl Methyl Trisulfide |
-3.4 |
-3.6 |
|
Diallyl Trisulfide |
-3.9 |
-4.0 |
|
S-Allylcystein |
-5.0 |
-4.6 |
|
Organosulfur |
-3.65 |
-4.37 |
|
Flavonoid |
|
|
|
Apigenin |
-8.1 |
-8.2 |
|
Myricetin |
-7.0 |
-8.0 |
|
Rutin |
-8.6 |
-7.5 |
|
Kaempferol |
-6.9 |
-8.2 |
|
Quercetin |
-7.8 |
-8.0 |
|
Isoquercitrin |
-8.9 |
-8.0 |
|
Flavonoid |
-7.8 |
-7.98 |
|
Terpenoid |
|
|
|
Citral |
-5.9 |
-5.7 |
|
Geraniol |
-5.8 |
-5.6 |
|
Linalool |
-5.7 |
-5.5 |
|
Caryophyllene |
-6.7 |
-7.1 |
|
α-Pinene |
-6.4 |
-5.4 |
|
Lupeol |
-8.7 |
-8.7 |
|
Terpenoid |
-6.53 |
-6.33 |
|
Saponins |
|
|
|
Agigenin |
-7.7 |
-8.2 |
|
Tigogenin |
-9.4 |
-6.5 |
|
Isoeruboside |
-8.6 |
-8.3 |
|
Sativoside B1 |
-8.3 |
-7.0 |
|
Sativoside R1 |
-7.8 |
-7.2 |
|
Sativoside R2 |
-8.3 |
-8.5 |
|
Saponins |
-8.35 |
-7.61 |
The docking study results between garlic active compounds with CCR5 and CXCR4 revealed that organosulfur family compounds had higher binding affinity values than the flavonoid, terpenoid, and saponin family groups (Table 2). CCR5-ligand docking (Figure 1) showed that the binding affinity value of S-allylcystein was the lowest among all the organosulfur compounds (–5.0 kcal/mol), but it still higher than isoquercitrin from flavonoid group, lupeol from terpenoid group, and tigogenin from saponins group (–8.9, –8.7, and –9.4 kcal/mol). A lower binding affinity value indicated that the binding of the protein-ligand was more stable. This docking study indicated that saponins are more effective as CCR5 inhibitors than organosulfur family groups.
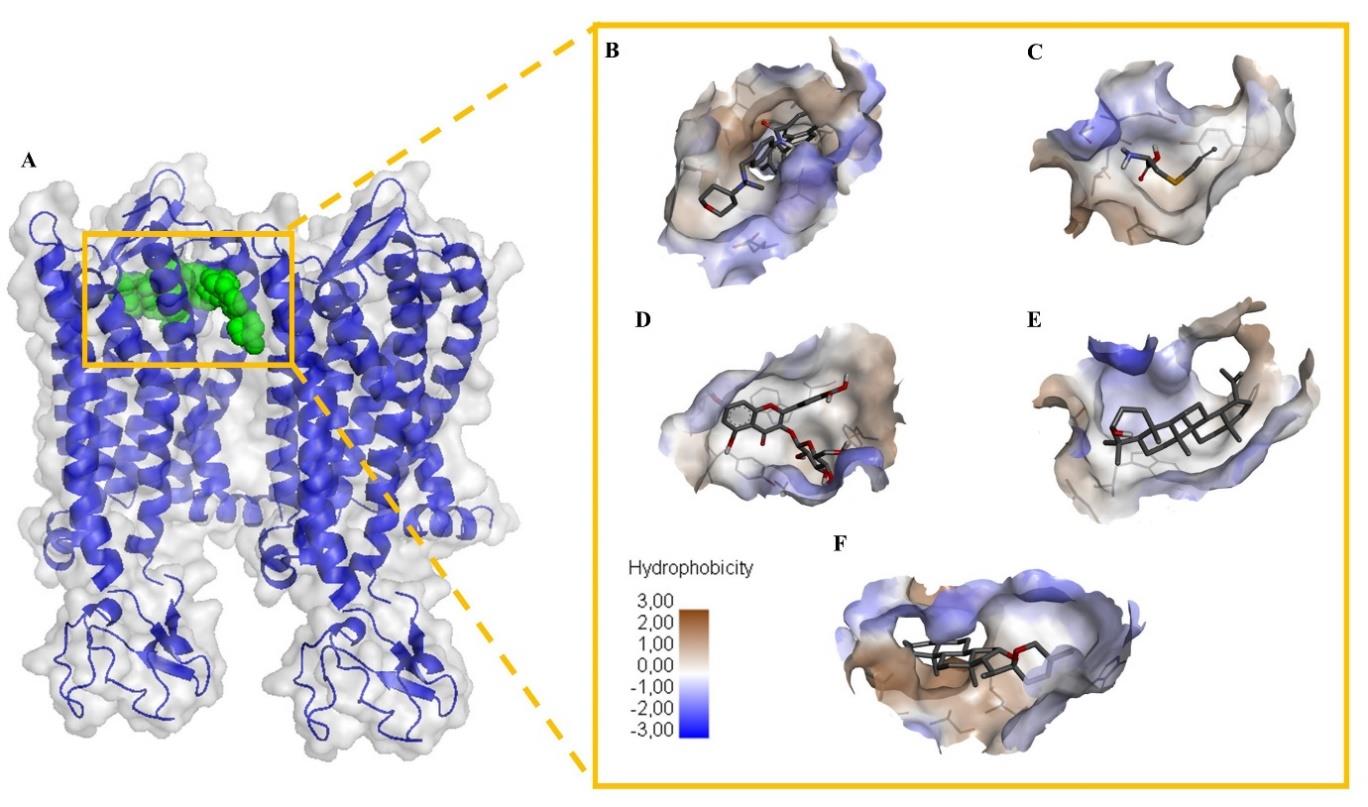
Figure 1. Visualization of molecular docking between active compounds found in garlic and protein CCR5. (A) Position of the ligand in the active site of the protein. The green sphere as ligand and surface blue cartoon as protein; docking of (B) CCR5 inhibitor known as TAK-9 (C) S-allylcystein (D) isoquercitrin (E) lupeol and (F) tigogenin. The yellow box contained interaction between compounds and CCR5 based on the hydrophobicity on the protein surface and amino acid residues for each compound. Visualization presented based on the lowest binding affinity value for each family compound in CCR5 docking. CCR5, C-C chemokine receptor type 5.
On the other hand, docking of the CXCR4-ligand (Figure 2) resulted in lupeol as one of the terpenoid family groups with the lowest binding affinity value (–8.7 kcal/mol). The binding affinity value of lupeol was also lower than that of the CXCR4 inhibitor (–8.6 kcal/mol), which indicates that lupeol could be more effective as a CXCR4 inhibitor. Meanwhile, molecular docking in CXCR4 with an organosulfur family group showed a higher binding affinity value than that of the non-organosulfur compound. Organosulfur compounds might be less stable when bound to CCR5 and CXCR4 (Table 2). rather than non-organosulfur compounds.
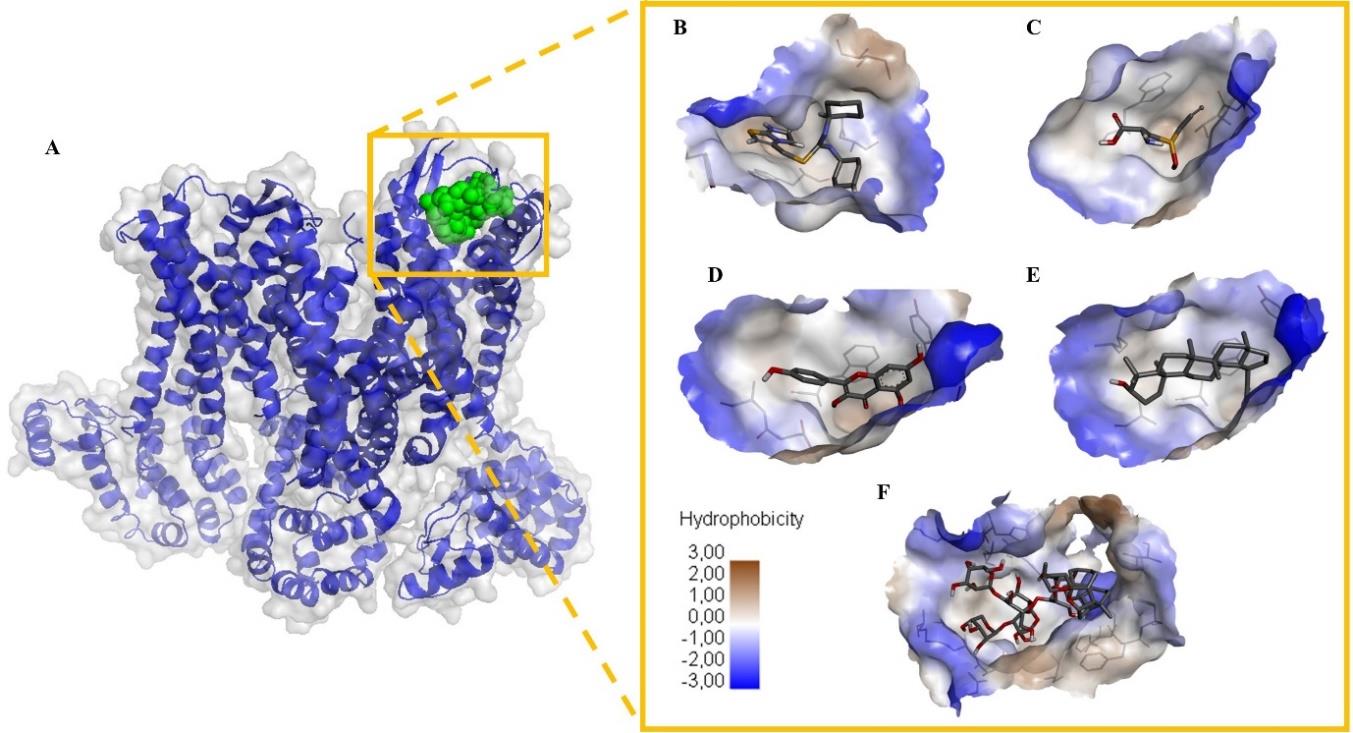
Figure 2. Visualization of molecular docking between active compounds found in garlic and protein CXCR4. (A) Position of the ligand in the active site of the protein. The green sphere as ligand and surface blue cartoon as protein; docking of (B) CXCR4 inhibitor known as IT1t (C) alliin (D) kaempferol (E) lupeol and (F) sativoside R2. Yellow box contained interaction between compounds and CXCR4 based on the hydrophobicity on the protein surface and amino acid residues for each compound. Visualization presented based on the lowest binding affinity value for each family compound in CXCR4 docking. CXCR4, C-X-C chemokine receptor type 4.
Protein interactions and networks analysis
Protein interaction analysis from BioGRID revealed CCR5 interactions with other proteins (Figure 3), especially evidence of the interaction between CCR5 and CXCR4 (Table 3). More than 15 proteins interact with CCR5. Based on BioGRID analysis showed that CCR5 and CXCR4 were involved in various cancer signaling pathways together with the Janus kinase (JAK), signal transducer and activator of transcription (STAT), JAK2, STA3, and STAT5 pathways.
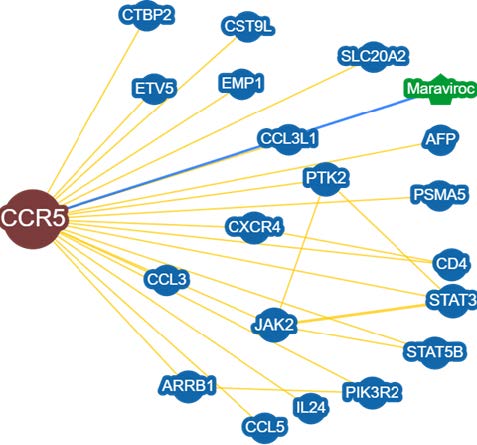
Figure 3. Protein interactions provided by BioGRID database.
Table 3. Protein interactions and evidence of interaction with CCR5.
|
Protein |
Evidence |
References |
|
AFP |
Affinity Capture-Western |
(Atemezem et al., 2002) |
|
ARRB1 |
Reconstituted Complex |
(Huttenrauch et al., 2002) |
|
CCL3 |
Reconstituted Complex |
(Navenot et al., 2001) |
|
CCL3L1 |
Reconstituted Complex |
(Miyakawa et al., 2002; Struyf et al., 2001) |
|
CCL5 |
Reconstituted Complex Affinity Capture-Western |
(Proudfoot et al., 2001; Struyf et al., 2001) (Slimani et al., 2003) |
|
CD4 |
Affinity Capture-Western |
(Xiao et al., 2000) |
|
CST9L |
Two-Hybrid |
(Wang et al., 2011) |
|
CTBP2 |
Two-Hybrid |
(Wang et al., 2011) |
|
CXCR4 |
Reconstituted Complex |
(Agrawal et al., 2002) |
|
EMP1 |
Two-Hybrid |
(Luck et al., 2020) |
|
ETV5 |
Two-Hybrid |
(Wang et al., 2011) |
|
IL24 |
Two-Hybrid |
(Wang et al., 2011) |
|
JAK2 |
Affinity Capture-Western |
(Wong et al., 2001) |
|
PIK3R2 |
Affinity Capture-Western |
(Mellado et al., 2001) |
|
PSMA5 |
Two-Hybrid Affinity Capture-Western |
(Fernandis et al., 2002) |
|
PTK2 |
Affinity Capture-Western |
(Cicala et al., 1999) |
|
SLC20A2 |
Two-Hybrid |
(Sokolina et al., 2017) |
|
STAT3 |
Affinity Capture-Western |
(Mellado et al., 2001) |
|
STAT5B |
Affinity Capture-Western |
(Mellado et al., 2001) |
Further analysis using the String Database revealed that CXCR4 is involved in the cancer pathway (red bulb) and interacts with CCR5 in the chemokine signaling pathway. Chemokines and their receptors have been reported that play both anticancer and pro-cancer roles. The CCR5 and CXCR4 pathways are involved in the development of cancer (Weitzenfeld and Ben-Baruch, 2014b). The inhibitory mechanisms of these chemokine receptors might have implications for cell growth in cancer progression.
DISCUSSION
Research on the discovery of novel compounds as anticancer candidates is promising and challenging. CCR5 and CXCR4 inhibitors may be promising candidates for cancer therapy based on their critical function during cancer progression (Weitzenfeld and Ben-Baruch, 2014a). Garlic possesses anticancer properties and regulates the phosphoinositide 3-kinase/serine-threonine kinase (PI3K/Akt) and JNK, B cell lymphoma-2 (Bcl2), and caspase 3 signaling pathways to induce apoptosis (Wang et al., 2010; Shin et al., 2014). However, its upstream mechanism of action, which involves the main receptor in cancer, remains unclear. Moreover, most garlic utilization is focused on organosulfur compounds. Herein, we compared the anticancer mechanisms of organosulfur and non-organosulfur compounds from garlic (Table 2) using an in-silico approach.
Surprisingly, the organosulfur compounds showed weaker binding capability than non-organosulfur compounds with CCR5 or CXCR4 (Table 2). Our molecular docking results demonstrated that tigogenin from the saponin group had a lower binding affinity than other compounds (except the CCR5 inhibitor or IT1t). Gibb's free energy measures the amount of free energy generated from the binding affinity. The more negative the Gibb's free energy value, the greater the interaction between the ligand and the protein (Atho’illah et al., 2021; Pertami et al., 2021). The key residues of CCR5 are Tyr37 and Trp86, with the latter being an important residue for the inhibition of CCR5 (Lin et al., 2019). Interestingly, our present study demonstrated that both organosulfur and non-organosulfur compounds have the same binding site as the CCR5 inhibitor at Tyr37, Trp86, and Gln280 (Figure 4). Tigogenin interacts with CCR5 through van der Waals interactions (Supplementary Table 1), which provides a major driving force for the binding of CCR5 (Lin et al., 2019). Meanwhile, another study reported that Gln280 plays a key role in the intermolecular interaction between CCL5 and CCR5, and the mutation of Gln280 significantly reduced the activity of CCR5 significantly (Tamamis and Floudas, 2014a, 2014b). The CCR5 binding cavity is mainly hydrophobic as various non-polar amino acids, including Trp86, influence the ligand-dependent CCR5 mechanism (Salmas et al., 2015). In cancer progression, CCR5 manipulates the chemokine networks to support tumor growth by leading them to the metastatic regions of cancer cell homing and increases the proinflammatory pro-metastatic immune phenotype, accelerates the DNA repair, ensures the survival of aberrant cells, and increases the resistance to DNA damage (Aldinucci et al., 2020). Therefore, blockade of the CCL5/CCR5 axis by counteracting the garlic constituents may inhibit the progression of cancer.
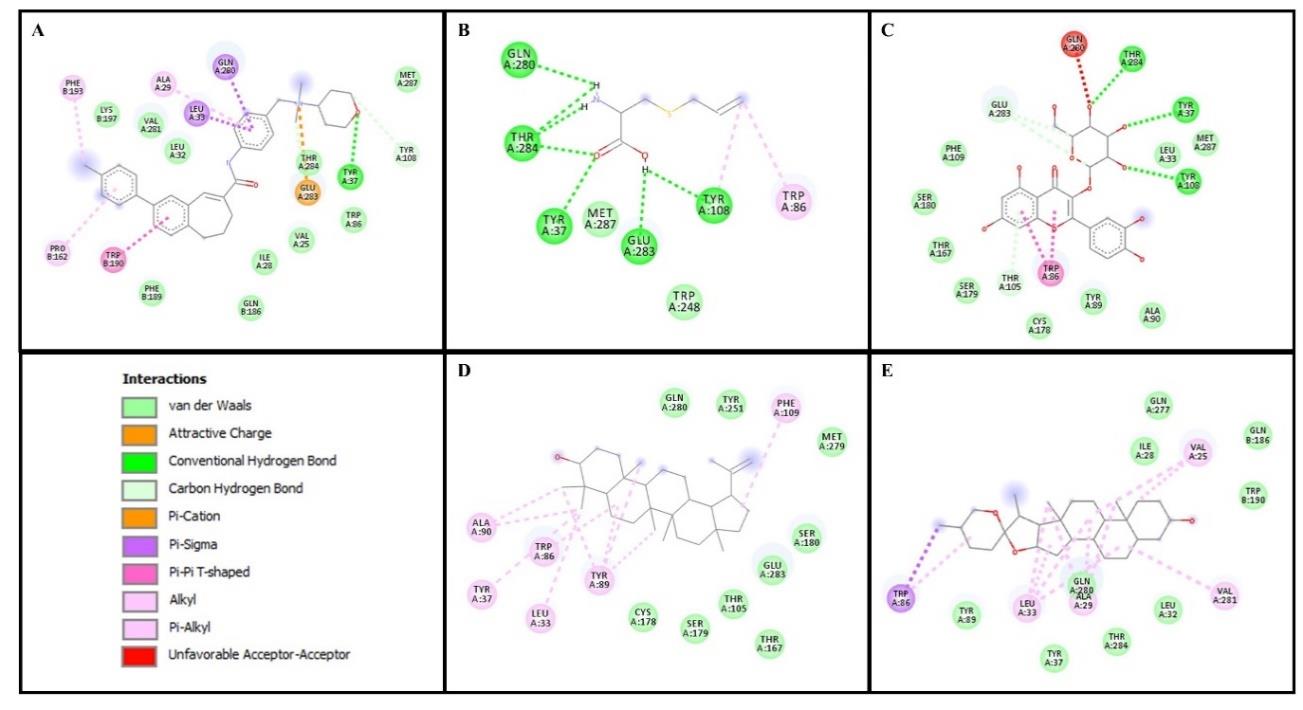
Figure 4. Amino acid residues and interaction types from molecular docking between CCR5 and ligands. (A) CCR5 inhibitor (TAK-79) (B) S-allylcystein (C) Isoquercitrin (D) lupeol and (E) tigogenin.
Furthermore, our present study demonstrated that lupeol from the terpenoid group showed the lowest binding affinity compared to other compounds (except the CXCR4 inhibitor) through van der Waals interactions (Supplementary Table 2). Interestingly, both organosulfur and non-organosulfur compounds have the same binding site as the CXCR4 inhibitor at Asp97, Leu41, Trp94, Ala98, and His113 (Figure 5). A previous study by Das et al. (2015) showed that these amino acid residues might have important roles in CXCR4, with Asp97 serving a critical role in cell fusion and is considered to be important for the binding of the CXCR4 inhibitor (Das et al., 2015). CXCR4 interaction with SDF-1 leads to the activation of several transduction signaling and their downstream pathways, which are involved in cell survival, proliferation, migration, adhesion, and chemotaxis (Xu et al., 2015). Notably, CXCR4 has either negligible or absent expression in normal cells, while it is overexpressed in the tumor cells close to normal cells (Chen et al., 2014). As mentioned earlier, the CXCR4/SDF-1 axis promotes tumor growth and attracts immune cells to the tumor sites; therefore, the blockade of this axis may ameliorate cancer metastasis (Mishra et al., 2016; Song et al., 2021). Our molecular docking results suggest that garlic compounds are bound to several key amino acids of CXCR4 and may have a beneficial impact in delaying the CXCR4 downstream mechanism during cancer progression.
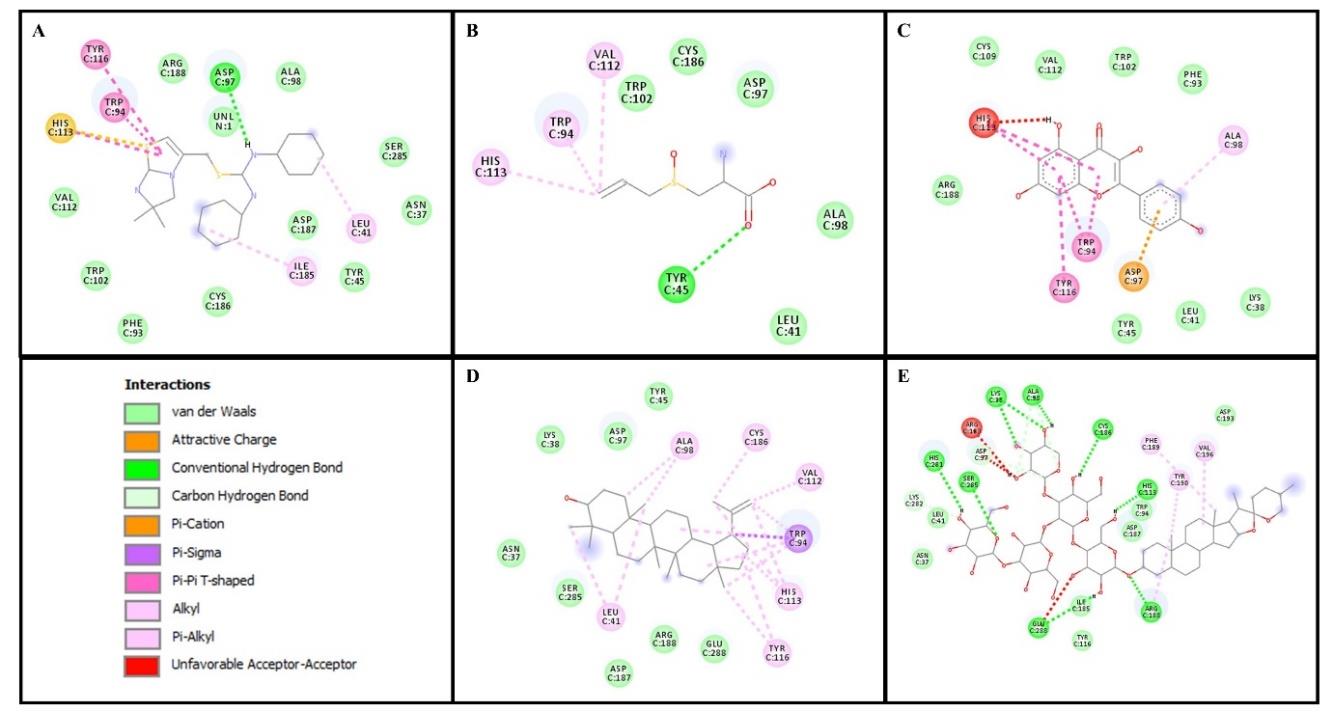
Figure 5. Amino acid residue and interaction from molecular docking between CXCR4 and ligands. (A) CXCR4 inhibitor (IT1t) (B) alliin (C) kaempferol (D) lupeol and (F) Sativoside R2.
The research study showed that all garlic compounds including organosulfur, terpenoids, flavonoids, and saponins groups can attach to target proteins with varying binding affinity values. Garlic is rich of organosulfur compounds (OSC) which classified into water-soluble organosulfur and oil-soluble organosulfur. Even though the mechanism of organosulfur compounds as anticancer is still unclear, previous research revealed that sulfur acts as a component in some amino acids and Fe-S clusters for enzyme activity. As we know, Fe-s plays an important role for origin of life such as DNA, RNA, and acetyl-CoA (Zeng et al., 2017). Omar & Al-Wabel (2010) reported that organosulfur compounds are involved in several metabolizing enzymes due to their structure, which act as activate or detoxify in cancer signaling. These findings suggested that organosulfur compounds might be involved as anticancer by modulating the activity of both of CCR5 and CXCR4 in cancer progression.
Free radicals are unstable molecules, very reactive, and cause DNA damage. Radicals-linked damage on DNA and proteins plays a significant role in cancer development (Pourahmad et al., 2016). Flavonoids are phytochemical compounds found in plants and polyphenols with diphenylpropane (C6-C3-C6) skeleton (Pasetto et al., 2014). Flavonoids characterized by the existence of functional hydroxyl groups. The structures mediate antioxidant activity by scavenging free radicals and/or by chelating metal ions. These functional hydroxyl groups especially the B ring hydroxyl configuration could be radical scavengers by donating the hydrogen and an electron to hydroxyl, peroxyl, and peroxynitrite radicals by forming flavonoids radical which are relatively more stable (Kumar & Pandey, 2013). On the other hand, flavonoids group has been reported of their ability to inhibit the human immunodeficiency virus (HIV) by affecting cluster of differentiation 4 (CD4) and co-receptors CCR5 and CXCR4 (Pasetto et al., 2014). The inhibitory activity of flavonoids over CCR5 and CXCR4 in HIV suggested that flavonoids might be able as an anticancer due to the inhibition role over CCR5 and CXCR4 in cancer progression.
Furthermore, another plant polyphenol which have antioxidant activity is terpenoids. Terpenoids consist with hydrocarbon chain made up of isoprene structural units. Monoterpenoid (C10), sesquiterpenoid (C15), diterpenoid (C20), triterpenoid (C30), tetraterpenoid (C40), polyterpenoid (C>40), etc. In addition to terpene hydrocarbons, terpenoids are synthesized through mevalonic acid (MVA) and 2C-methyl-D-erythritol-4-phosphate (MEP) biochemical pathway. Both pathways provide terpenes skeletons, which results in functionalized and physiologically active secondary metabolites (Bergman et al., 2019). For example, geraniol induces tumor cell apoptosis and inhibits Akt signaling pathway exhibit an anticancer effect (Kim et al., 2011). Interestingly, lupeol which showed the strongest inhibitory towards CXCR4 could act as multi-target agent which involve in the several important pathways, including PI3K/Akt signaling pathway (Saleem, 2009; Liu et al., 2013). Another study reported that the isolated terpenoid inhibit JAK2 phosphorylation through interaction with FERM-SH2 domain of JAK2 resulting in STAT3 inhibition (Bailly, 2020). These effects are necessary to delay cancer progression, which further supported by our networking analysis.
Garlic also contains with high levels of terpenoids, flavonoids, steroidal saponins, and so on. Steroidal saponins are widely distributed among monocotyledonous families. Saponin is structurally made up from distinct components consisting of an isoprenoid unit and sugar residue oligosaccharides as glycone part. Saponins group especially steroidal saponins contain aglycone part at the former, while glycone at the end of chain. Saponins consist of a hydrophilic sugar moiety and a hydrophobic genin called sapogenin (Elekofehinti et al., 2021). The steroidal sapogenins contain a cholestane nucleus, and side-chain cyclization may occur, yielding furostanes, furosprostanes, and spirostanes. Tigogenin, a spirostane-tipe sapogenin, which possess both tetrahydropyran and tetrahydrofuran rings joined at C-22 bearing saccharides chains displayed antitumor activities in cancer several cell lines (Simmons-Boyce and Tinto, 2007; Juang and Liang, 2020).
In addition, our network analysis revealed that both CCR5 and CXCR4 share the same pathway via the cancer pathway, chemokine signaling pathway, and immune system (Figure. 6). These three pathways play pivotal roles in cancer progression. CCR5 is responsible for the establishment of the cancer microenvironment in various cancer types. CCR5 manipulates the immune cells to become a pro-metastatic immune phenotype to build a protective microenvironment, leading to cancer cell growth, survival, migration, and invasion during metastasis (Jiao et al., 2019; Aldinucci et al., 2020; Pervaiz et al., 2021). CXCR4 regulates cell division via JAK2/3 in the PI3K/Akt signaling pathway (Abdelouahab et al., 2017). Targeting CCR5 and CXCR4 upstream in the cancer pathway might be an effective strategy for treating cancer.
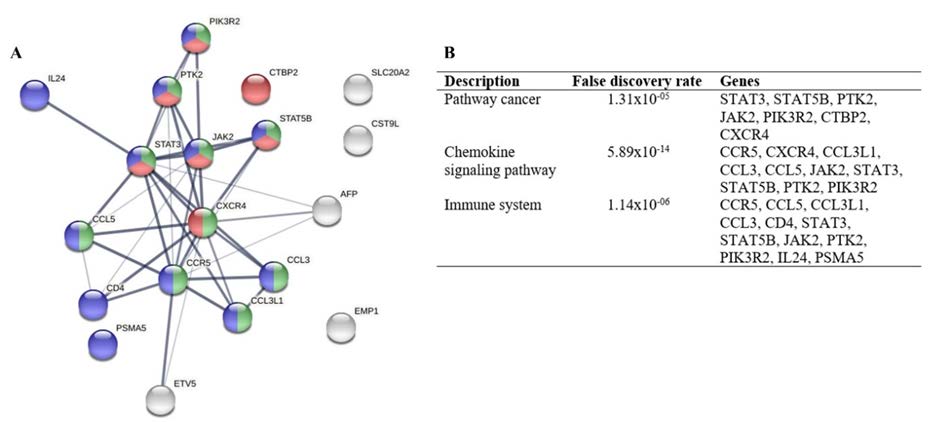
Figure 6. Result of CCR5 and CXCR4 networks. (A) Protein networks based on String Database, Pathway cancer was marked with red color, Chemokine signaling pathway with green color, and Immune system with blue color. (B) False discovery and identification of genes that engaged for each pathway. CCR5, C-C chemokine receptor type 5; CXCR4, C-X-C chemokine receptor type 4.
Therefore, organosulfur compounds have been reported to block the CCR5 and CXCR4 signaling pathways, leading to apoptosis and delayed cancer proliferation (Wang et al., 2010; Shin et al., 2014; Guan et al., 2015; Song et al., 2015; Lee et al., 2018). A previous study reported that tigogenin has a weak anti-proliferative activity; in contrast, tigogenin synthesis to its analogs or derivatives might transform tigogenin to be a more potent and selective anticancer candidate (Li et al., 2018; Michalak et al., 2020). Numerous studies have shown that lupeol induces apoptosis in cancer cells by inhibiting the PI3K/Akt pathway and suppressing STAT3 activity. In this scenario, based on our molecular docking results, the non-organosulfur in garlic was identified as a promising candidate to interfere with CCR5 and CXCR4 signaling pathways for improving the treatment to treat or possibly improve the quality of life of cancer patients.
CONCLUSION
The results of the present study suggest that the non-organosulfur compounds from garlic might act as potential CCR5 or CXCR4 inhibitors, with better efficacies than the organosulfur compounds. Tigogenin from the saponin group was predicted to be a CCR5 inhibitor, while lupeol from the terpenoid group was predicted to be a CXCR4 inhibitor. These garlic compounds may be used to regulate both the CCR5 and CXCR4 signaling pathways to inhibit the proliferation, migration, and survival of cancer cells. However, further in vitro and in vivo studies should be conducted to validate the activities of garlic compounds and determine their efficacies to be used as primary compounds for the prevention of cancer progression.
ACKNOWLEDGEMENTS
Authors gratitude is addressed to Assoc. Prof. Dr. Sri Rahayu Lestari, M.Si for the helpful discussion.
AUTHOR CONTRIBUTIONS
Balqis Balqis designed and conducting the experiments, perform analysis and interpretation, and wrote the manuscript. Betty Lukiati and Mohamad Amin assisted in conducting the experiments and make critical review. Siti Nur Arifah and Mochammad Fitri Atho’illah assisted in data collection and processing, data analysis or interpretation, and writing manuscript. Nashi Widodo designed and supervised all of the experiments and make critical review of the manuscript. All authors have read and approved of the final manuscript.
CONFLICT OF INTEREST
The authors declare that they hold no competing interests.
REFERENCES
Abdelouahab, H., Zhang, Y., Wittner, M., Oishi, S., Fujii, N., Besancenot, R., Plo, I., Ribrag, V., Solary, E., Vainchenker, W., et al. 2017. CXCL12/CXCR4 pathway is activated by oncogenic JAK2 in a PI3K-dependent manner. Oncotarget. 8: 54082–54095.
Agrawal, L., Vanhorn-Ali, Z., and Alkhatib, G. 2002. Multiple determinants are involved in HIV coreceptor use as demonstrated by CCR4/CCL22 interaction in peripheral blood mononuclear cells (PBMCs). Journal of Leukocyte Biology. 72: 1063–1074.
Aldinucci, D., Borghese, C., and Casagrande, N. 2020. The CCL5/CCR5 Axis in Cancer Progression. Cancers. 12: 1765.
Allemani, C. and Coleman, M.P. 2017. Public health surveillance of cancer survival in the United States and worldwide: The contribution of the CONCORD programme: Public Health Surveillance of Cancer Survival. Cancer. 123: 4977–4981.
Amagase, H. 2006. Significance of Garlic and Its Constituents in Cancer and Cardiovascular Disease. The Journal of Nutrition. 136: 716S-725S.
Arifah, S.N., Atho’illah, M.F., Lukiati, B., and Lestari, S.R. 2020. Herbal Medicine from Single Clove Garlic Oil Extract Ameliorates Hepatic Steatosis and Oxidative Status in High Fat Diet Mice. The Malaysian Journal of Medical Sciences: MJMS, 27: 46–56.
Arimont, M., Sun, S.-L., Leurs, R., Smit, M., de Esch, I.J.P., and de Graaf, C. 2017. Structural Analysis of Chemokine Receptor–Ligand Interactions. Journal of Medicinal Chemistry. 60: 4735–4779.
Atemezem, A., Mbemba, E., Marfaing, R., Vaysse, J., Pontet, M., Saffar, L., Charnaux, N., and Gattegno, L. 2002. Human alpha-fetoprotein binds to primary macrophages. Biochemical and Biophysical Research Communications. 296: 507–514.
Atho’illah, M.F., Safitri, Y.D., Nur’aini, F.D., Widyarti, S., Tsuboi, H., and Rifa’i, M. 2021. Elicited soybean extract attenuates proinflammatory cytokines expression by modulating TLR3/TLR4 activation in high-fat, high-fructose diet mice. Journal of Ayurveda and Integrative Medicine. 12: 43–51.
Bailly, C. 2020. Anticancer activities and mechanism of action of nagilactones, a group of terpenoid lactones isolated from podocarpus species. Natural Products and Bioprospecting. 10: 367–375.
Balqis, Widodo, Lukiati, B., and Amin, M. 2018. Active compounds with antioxidant potential in boiled local Papua-Indonesian garlic. 020012.
Bergman, M.E., Davis, B., and Phillips, M.A. 2019. Medically Useful Plant Terpenoids: Biosynthesis, Occurrence, and Mechanism of Action. Molecules, 24: 3961.
Bohn, O.L., Nasir, I., Brufsky, A., Tseng, G.C., Bhargava, R., MacManus, K., and Chivukula, M. 2009. Biomarker profile in breast carcinomas presenting with bone metastasis. International Journal of Clinical and Experimental Pathology. 3: 139–146.
Cao, H.-X., Zhu, K.-X., Fan, J.-G., and Qiao, L. 2014. Garlic-derived allyl sulfides in cancer therapy. Anticancer Agents in Medicinal Chemistry. 14: 793–799.
Chan, K.K.L., Yao, T.J., Jones, B., Zhao, J.F., Ma, F.K., Leung, C.Y., Lau, S.K., Yip, M.W., and Ngan, H.Y.S. 2011. The use of Chinese herbal medicine to improve quality of life in women undergoing chemotherapy for ovarian cancer: A double-blind placebo-controlled randomized trial with immunological monitoring. Annals of Oncology. 22: 2241–2249.
Chen, Y., Wei, Y., Liu, J., and Zhang, H. 2014. Chemotactic responses of neural stem cells to SDF-1α correlate closely with their differentiation status. Journal of Molecular Neuroscience. 54: 219–233.
Cheng, Y.-Y., Hsieh, C.-H.and Tsai, T.-H. 2018. Concurrent administration of anticancer chemotherapy drug and herbal medicine on the perspective of pharmacokinetics. Journal of Food and Drug Analysis. 26: S88–S95.
Cicala, C., Arthos, J., Ruiz, M., Vaccarezza, M., Rubbert, A., Riva, A., Wildt, K., Cohen, O., and Fauci, A.S. 1999. Induction of phosphorylation and intracellular association of CC chemokine receptor 5 and focal adhesion kinase in primary human CD4+ T cells by macrophage-tropic HIV envelope. Journal of Immunology (Baltimore, Md.: 1950). 163: 420–426.
Dallakyan, S. and Olson, A.J. 2015. Small-molecule library screening by docking with PyRx. Methods in Molecular Biology (Clifton, N.J.). 1263: 243–250.
Das, D., Maeda, K., Hayashi, Y., Gavande, N., Desai, D.V., Chang, S.B., Ghosh, A.K., and Mitsuya, H. 2015. Insights into the Mechanism of Inhibition of CXCR4: Identification of Piperidinylethanamine Analogs as Anti-HIV-1 Inhibitors. Antimicrobial Agents and Chemotherapy. 59: 1895–1904.
Dassault Systèmes BIOVIA. 2015. Discovery studio modeling environment, Version 4.5. Dassault Systèmes.
Debnath, B., Xu, S., Grande, F., Garofalo, A., and Neamati, N. 2013. Small Molecule Inhibitors of CXCR4. Theranostics. 3: 47–75.
Du, X., Li, Y., Xia, Y.-L., Ai, S.-M., Liang, J., Sang, P., Ji, X.-L., and Liu, S.-Q. 2016. Insights into protein–ligand interactions: mechanisms, models, and methods. International Journal of Molecular Sciences. 17.
Elekofehinti, O.O., Iwaloye, O., Olawale, F., and Ariyo, E.O. 2021. Saponins in cancer treatment: current progress and future prospects. Pathophysiology, 28: 250–272.
Fernandis, A.Z., Cherla, R.P., Chernock, R.D., and Ganju, R.K. 2002. CXCR4/CCR5 down-modulation and chemotaxis are regulated by the proteasome pathway. Journal of Biological Chemistry. 277: 18111–18117.
García-Cuesta, E.M., Santiago, C.A., Vallejo-Díaz, J., Juarranz, Y., Rodríguez-Frade, J.M., and Mellado, M. 2019. The Role of the CXCL12/CXCR4/ACKR3 axis in autoimmune diseases. Frontiers in Endocrinology. 0.
Gilson, M.K., and Zhou, H.-X. 2007. Calculation of protein-ligand binding affinities. Annual Review of Biophysics and Biomolecular Structure. 36: 21–42.
Global Burden of Disease Cancer Collaboration, Fitzmaurice, C., Akinyemiju, T.F., Al Lami, F.H., Alam, T., Alizadeh-Navaei, R., Allen, C., Alsharif, U., Alvis-Guzman, N., Amini, E., et al. 2018. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups, 1990 to 2016: a systematic analysis for the global burden of disease study. JAMA Oncology, 4: 1553.
Guan, G., Zhang, Y., Lu, Y., Liu, L., Shi, D., Wen, Y., Yang, L., Ma, Q., Liu, T., Zhu, X., Qiu, X., and Zhou, Y. 2015. The HIF-1α/CXCR4 pathway supports hypoxia-induced metastasis of human osteosarcoma cells. Cancer Letters. 357: 254–264.
Heron, M., and Anderson, R.N. 2016. Changes in the leading cause of death: recent patterns in heart disease and cancer mortality. NCHS Data Brief. 254: 1–8.
Hu, X.-Q., Sun, Y., Lau, E., Zhao, M., and Su, S.-B. 2016. Advances in synergistic combinations of chinese herbal medicine for the treatment of cancer. Current Cancer Drug Targets. 16: 346–356.
Huang, H., Zhang, G., Zhou, Y., Lin, C., Chen, S., Lin, Y., Mai, S., and Huang, Z. 2018. Reverse Screening Methods to Search for the Protein Targets of Chemopreventive Compounds. Frontiers in Chemistry. 6: 138.
Huttenrauch, F., Nitzki, A., Lin, F.-T., Höning, S., and Oppermann, M. 2002. Beta-arrestin binding to CC chemokine receptor 5 requires multiple C-terminal receptor phosphorylation sites and involves a conserved Asp-Arg-Tyr sequence motif. Journal of Biological Chemistry. 277: 30769–30777.
Jermini, M., Dubois, J., Rodondi, P.-Y., Zaman, K., Buclin, T., Csajka, C., Orcurto, A., and Rothuizen, L.E. 2019. Complementary medicine use during cancer treatment and potential herb-drug interactions from a cross-sectional study in an academic centre. Scientific Reports. 9: 5078.
Jiao, X., Nawab, O., Patel, T., Kossenkov, A.V., Halama, N., Jaeger, D., and Pestell, R.G. 2019. Recent advances targeting CCR5 for cancer and its role in immuno-oncology. Cancer Research. 79: 4801–4807.
Juang, Y.-P. and Liang, P.-H. 2020. Biological and Pharmacological effects of synthetic saponins. Molecules. 25: 4974.
Kalpana, G., Figy, C., Yeung, M., and Yeung, K.C. 2019. Reduced RhoA expression enhances breast cancer metastasis with a concomitant increase in CCR5 and CXCR4 chemokines signaling. Scientific Reports. 9: 16351.
Kim, S.-H., Park, E.-J., Lee, C.-R., Chun, J. N., Cho, N.-H., Kim, I.-G., Lee, S.-H., Kim, T.-W., Park, H.-H., So, I.-S., et al. 2011. Geraniol induces cooperative interaction of apoptosis and autophagy to elicit cell death in PC-3 prostate cancer cells. International Journal of Oncology.
Kumar, S. and Pandey, A.K. 2013. Chemistry and Biological Activities of Flavonoids: An Overview. The Scientific World Journal. 2013: e162750.
Lau, G., Labrecque, J., Metz, M., Vaz, R., and Fricker, S.P. 2015. Specificity for a CCR5 Inhibitor Is Conferred by a Single Amino Acid Residue: ROLE OF ILE198. Journal of Biological Chemistry. 290: 11041–11051.
Lee, H.H., Jeong, J.-W., Hong, S.H., Park, C., Kim, B.W., and Choi, Y.H. 2018. Diallyl trisulfide suppresses the production of lipopolysaccharide-induced inflammatory mediators in BV2 microglia by decreasing the NF-κB pathway activity associated with toll-like receptor 4 and CXCL12/CXCR4 pathway blockade. Journal of Cancer Prevention. 23: 134–140.
Lestari, S.R., Atho’illah, M.F., Christina, Y.I., and Rifa’i, M. 2020. Single garlic oil modulates T cells activation and proinflammatory cytokine in mice with high fat diet. Journal of Ayurveda and Integrative Medicine. 11: 414–420.
Lestari, S.R. and Rifai, M. 2019. The effect of single-bulb garlic oil extract toward the hematology and histopathology of the liver and kidney in mice. Brazilian Journal of Pharmaceutical Sciences. 55: e18027.
Lestari, S.R. and Rifa’i, M. 2019. Regulatory T cells and anti-inflammatory cytokine profile of mice fed a high-fat diet after single-bulb garlic (Allium sativum L.) oil treatment. Tropical Journal of Pharmaceutical Research. 17: 2157.
Li, G.-L., Xu, H.-J., Xu, S.-H., Wang, W.-W., Yu, B.-Y., and Zhang, J. 2018. Synthesis of tigogenin MeON-Neoglycosides and their antitumor activity. Fitoterapia. 125: 33–40.
Lin, H.-Y., Ho, Y., and Liu, H.-L. 2019. Structure-based pharmacophore modeling to discover novel CCR5 inhibitors for HIV-1/cancers therapy. Journal of Biomedical Science and Engineering. 12: 10–30.
Liu, F., He, Y., Liang, Y., Wen, L., Zhu, Y., Wu, Y., Zhao, L., Li, Y., Mao, X., and Liu, H. 2013. PI3-kinase inhibition synergistically promoted the anti-tumor effect of lupeol in hepatocellular carcinoma. Cancer Cell International. 13: 108.
Luck, K., Kim, D.-K., Lambourne, L., Spirohn, K., Begg, B.E., Bian, W., Brignall, R., Cafarelli, T., Campos-Laborie, F.J., Charloteaux, B., et al. 2020. A reference map of the human binary protein interactome. Nature. 580: 402–408.
Mellado, M., Rodríguez-Frade, J.M., Vila-Coro, A.J., Fernández, S., Martín de Ana, A., Jones, D.R., Torán, J.L., and Martínez-A, C. 2001. Chemokine receptor homo- or heterodimerization activates distinct signaling pathways. The EMBO Journal. 20: 2497–2507.
Michalak, O., Krzeczyński, P., Cieślak, M., Cmoch, P., Cybulski, M., Królewska-Golińska, K., Kaźmierczak-Barańska, J., Trzaskowski, B., and Ostrowska, K. 2020. Synthesis and anti–tumour, immunomodulating activity of diosgenin and tigogenin conjugates. The Journal of Steroid Biochemistry and Molecular Biology, 198, 105573.
Mirzaei, H., Zarbafian, S., Villar, E., Mottarella, S., Beglov, D., Vajda, S., Paschalidis, I. Ch., Vakili, P., and Kozakov, D. 2015. Energy minimization on manifolds for docking flexible molecules. Journal of Chemical Theory and Computation. 11: 1063–1076.
Mishra, R.K., Shum, A.K., Platanias, L.C., Miller, R.J., and Schiltz, G.E. 2016. Discovery and characterization of novel small-molecule CXCR4 receptor agonists and antagonists. Scientific Reports. 6: 30155.
Miyakawa, T., Obaru, K., Maeda, K., Harada, S., and Mitsuya, H. 2002. Identification of amino acid residues critical for LD78beta, a variant of human macrophage inflammatory protein-1alpha, binding to CCR5 and inhibition of R5 human immunodeficiency virus type 1 replication. Journal of Biological Chemistry. 277: 4649–4655.
Navenot, J.M., Wang, Z.X., Trent, J.O., Murray, J.L., Hu, Q.X., DeLeeuw, L., Moore, P.S., Chang, Y., and Peiper, S.C. 2001. Molecular anatomy of CCR5 engagement by physiologic and viral chemokines and HIV-1 envelope glycoproteins: Differences in primary structural requirements for RANTES, MIP-1 alpha, and vMIP-II Binding. Journal of Molecular Biology. 313: 1181–1193.
O’Boyle, N.M., Banck, M., James, C.A., Morley, C., Vandermeersch, T., and Hutchison, G. R. 2011. Open Babel: An open chemical toolbox. Journal of Cheminformatics, 3: 33.
Omar, S.H. and Al-Wabel, N.A. 2010. Organosulfur compounds and possible mechanism of garlic in cancer. Saudi Pharmaceutical Journal. 18: 51–58.
Pasetto, S., Pardi, V., and Murata, R.M. 2014. Anti-HIV-1 activity of flavonoid myricetin on HIV-1 infection in a dual-chamber in vitro model. PLOS ONE. 9: e115323.
Pertami, S.B., Arifah, S.N., Atho’illah, M.F., and Budiono, B. 2021. Active compounds from polyscias scutellaria stimulate breast milk production: in silico study on serotonergic 5-HT2A receptors and prolactin receptors. Tropical Journal of Natural Product Research. 5: 1223–1229.
Pervaiz, A., Zepp, M., Georges, R., Bergmann, F., Mahmood, S., Faiza, S., Berger, M.R., & Adwan, H. 2021. Antineoplastic effects of targeting CCR5 and its therapeutic potential for colorectal cancer liver metastasis. Journal of Cancer Research and Clinical Oncology. 147: 73–91.
Pourahmad, J., Salimi, A., and Seydi, E. 2016. Role of Oxygen Free Radicals in Cancer Development and Treatment. In Free Radicals and Diseases. IntechOpen.
Proudfoot, A.E., Fritchley, S., Borlat, F., Shaw, J.P., Vilbois, F., Zwahlen, C., Trkola, A., Marchant, D., Clapham, P.R., and Wells, T.N. 2001. The BBXB motif of RANTES is the principal site for heparin binding and controls receptor selectivity. Journal of Biological Chemistry, 276: 10620–10626.
Saleem, M. 2009. Lupeol, a novel anti-inflammatory and anti-cancer dietary triterpene. Cancer Letters. 285: 109–115.
Salmas, R. E., Yurtsever, M., and Durdagi, S. 2015. Investigation of inhibition mechanism of chemokine receptor CCR5 by micro-second molecular dynamics simulations. Scientific Reports. 5: 13180.
Shin, D.Y., Kim, G.-Y., Hwang, H.J., Kim, W.-J., and Choi, Y.H. 2014. Diallyl trisulfide-induced apoptosis of bladder cancer cells is caspase-dependent and regulated by PI3K/Akt and JNK pathways. Environmental Toxicology and Pharmacology, 37: 74–83.
Simmons-Boyce, J.L., and Tinto, W.F. 2007. Steroidal saponins and sapogenins from the agavaceae family. Natural Product Communications. 2: 1934578X0700200.
Slimani, H., Charnaux, N., Mbemba, E., Saffar, L., Vassy, R., Vita, C., and Gattegno, L. 2003. Interaction of RANTES with syndecan-1 and syndecan-4 expressed by human primary macrophages. Biochimica Et Biophysica Acta. 1617: 80–88.
Smith, P.J., Clavarino, A., Long, J., and Steadman, K.J. 2014. Why do some cancer patients receiving chemotherapy choose to take complementary and alternative medicines and what are the risks?: CAM use by cancer patients. Asia-Pacific Journal of Clinical Oncology. 10: 1–10.
Sokolina, K., Kittanakom, S., Snider, J., Kotlyar, M., Maurice, P., Gandía, J., Benleulmi-Chaachoua, A., Tadagaki, K., Oishi, A., et al. 2017. Systematic protein-protein interaction mapping for clinically relevant human GPCRs. Molecular Systems Biology. 13: 918.
Song, B., Shu, Y., Cui, T., and Fu, P. 2015. Allicin inhibits human renal clear cell carcinoma progression via suppressing HIF pathway. International Journal of Clinical and Experimental Medicine. 8: 20573–20580.
Song, J.-S., Chang, C.-C., Wu, C.-H., Dinh, T.K., Jan, J.-J., Huang, K.-W., Chou, M.-C., Shiue, T.-Y., Yeh, K.-C., Ke, Y.-Y., et al. 2021. A highly selective and potent CXCR4 antagonist for hepatocellular carcinoma treatment. Proceedings of the National Academy of Sciences. 118: e2015433118.
Struyf, S., Menten, P., Lenaerts, J.P., Put, W., D’Haese, A., De Clercq, E., Schols, D., Proost, P., and Van Damme, J. 2001. Diverging binding capacities of natural LD78beta isoforms of macrophage inflammatory protein-1alpha to the CC chemokine receptors 1, 3 and 5 affect their anti-HIV-1 activity and chemotactic potencies for neutrophils and eosinophils. European Journal of Immunology. 31: 2170–2178.
Szklarczyk, D., Franceschini, A., Wyder, S., Forslund, K., Heller, D., Huerta-Cepas, J., Simonovic, M., Roth, A., Santos, A., et al. 2015. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Research. 43: D447-452.
Tamamis, P. and Floudas, C.A. 2014a. Molecular Recognition of CCR5 by an HIV-1 gp120 V3 Loop. PLoS ONE. 9: e95767.
Tamamis, P. and Floudas, C.A. 2014b. Elucidating a key anti-HIV-1 and cancer-associated axis: The structure of CCL5 (Rantes) in complex with CCR5. Scientific Reports. 4: 5447.
The Global Cancer Observatory. 2020. 360-Indonesia-Fact-Sheet. International Agency for Research on Cancer: World Health Organization. https://gco.iarc.fr/today/data/factsheets/populations/360-indonesia-fact-sheets.pdf
Torre, L.A., Siegel, R.L., Ward, E.M., and Jemal, A. 2016. Global Cancer Incidence and Mortality Rates and Trends—An Update. Cancer Epidemiology Biomarkers & Prevention. 25: 16–27.
Trott, O. and Olson, A.J. 2010. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. Journal of Computational Chemistry. 31: 455–461.
Varshney, R. and Budoff, M.J. 2016. Garlic and Heart Disease. Journal of Nutrition. 146: 416S-421S.
Wachtel-Galor, S. and Benzie, I.F.F. 2011. Herbal Medicine: An Introduction to Its History, Usage, Regulation, Current Trends, and Research Needs. In I.F.F. Benzie and S. Wachtel-Galor (Eds.), Herbal Medicine: Biomolecular and Clinical Aspects (2nd ed.). CRC Press/Taylor & Francis. http://www.ncbi.nlm.nih.gov/books/NBK92773/
Wang, J., He, L., Combs, C.A., Roderiquez, G., and Norcross, M.A. 2006. Dimerization of CXCR4 in living malignant cells: Control of cell migration by a synthetic peptide that reduces homologous CXCR4 interactions. Molecular Cancer Therapeutics. 5: 2474–2483.
Wang, J., Huo, K., Ma, L., Li, D., Huang, X., Yuan, Y., Li, C., Wang, Wei, Guan, W., et al. 2011. Toward an understanding of the protein interaction network of human liver. Molecular System Biology. 7: 536
Wang, Y.-B., Qin, J., Zheng, X.-Y., Bai, Y., Yang, K., and Xie, L.-P. 2010. Diallyl trisulfide induces Bcl-2 and caspase-3-dependent apoptosis via downregulation of Akt phosphorylation in human T24 bladder cancer cells. Phytomedicine. 17: 363–368.
Weitzenfeld, P. and Ben-Baruch, A. 2014. The chemokine system, and its CCR5 and CXCR4 receptors, as potential targets for personalized therapy in cancer. Cancer Letters. 352: 36–53.
Wong, M., Uddin, S., Majchrzak, B., Huynh, T., Proudfoot, A.E., Platanias, L.C., and Fish, E.N. 2001. Rantes activates Jak2 and Jak3 to regulate engagement of multiple signaling pathways in T cells. The Journal of Biological Chemistry. 276: 11427–11431.
World Health Organization. 2018. Latest global cancer data: Cancer burden rises to 18.1 million new cases and 9.6 million cancer deaths in 2018 (No. 263; pp. 1–3). https://www.who.int/cancer/PRGlobocanFinal.pdf
Wu, X., Chung, V.C.H., Lu, P., Poon, S.K., Hui, E.P., Lau, A.Y.L., Balneaves, L.G., Wong, S.Y.S., and Wu, J.C.Y. 2016. Chinese Herbal Medicine for Improving Quality of Life Among Nonsmall Cell Lung Cancer Patients: Overview of Systematic Reviews and Network Meta-Analysis. Medicine. 95: e2410.
Xiao, X., Kinter, A., Broder, C.C., and Dimitrov, D.S. 2000. Interactions of CCR5 and CXCR4 with CD4 and gp120 in human blood monocyte-derived dendritic cells. Experimental and Molecular Pathology. 68: 133–138.
Xu, C., Zhao, H., Chen, H., and Yao, Q. 2015. CXCR4 in breast cancer: Oncogenic role and therapeutic targeting. Drug Design, Development and Therapy. 4953.
Zeng, Y., Li, Y., Yang, J., Pu, X., Du, J., Yang, X., Yang, T., and Yang, S. 2017. Therapeutic Role of Functional Components in Alliums for Preventive Chronic Disease in Human Being. Evidence-Based Complementary and Alternative Medicine. 2017: e9402849.
Zhang, J., Wang, J., Qian, Z., and Han, Y. 2019. CCR5 is Associated with Immune Cell Infiltration and Prognosis of Lung Cancer. Journal of Thoracic Oncology. 14: e102–e103.
OPEN access freely available online
Chiang Mai University Journal of Natural Sciences [ISSN 16851994]
Chiang Mai University, Thailand
https://cmuj.cmu.ac.th
Balqis Balqis1, Betty Lukiati1, Mohamad Amin1, Siti Nur Arifah1, Mochammad Fitri Atho’illah2, and Nashi Widodo2,*
1 Department of Biology, Faculty of Mathematics and Sciences, Universitas Negeri Malang, 65145, Malang, East Java, Indonesia
2 Department of Biology, Faculty of Mathematics and Natural Sciences, Brawijaya University, 65145, Malang, East Java, Indonesia
Corresponding author: Nashi Widodo, E-mail: widodo@ub.ac.id
Total Article Views
Editor: Nisit Kittipongpatana,
Chiang Mai University, Thailand
Article history:
Received: Mach 15 2021;
Revised: October 5, 2021;
Accepted: October 21, 2021

