
Natural Bioactive Compounds of Sponge-Associated Fungi with Three Marine Ecosystems in Karimunjawa Island, Indonesia
Muhammad Zainuddin, Delianis Pringgenies*, Ocky Karna Radjasa and HaeruddinPublished Date : 2022-01-21
DOI : https://doi.org/10.12982/CMUJNS.2022.009
Journal Issues : Number 1, January-March 2022
Abstract Sponges are marine organisms that have associations with microorganisms. The association fungi in sponges have a chemical and ecological relationship. These bioactive compounds have potential in the pharmaceutical field. This research aimed to explore bioactive compounds of association fungi in sponges that live in coral, seaweed and mangrove ecosystems. The research consisted of isolation, purification, extraction, antibacterial testing, molecular identification, antioxidant testing and toxicity testing. The research was conducted on March to September 2020 in Karimunjawa Island. The results showed that the sponge association fungi isolates had antibacterial activity against the Multi Drug Resistant (MDR) pathogens Staphylococcus aureus (Sa), Escherichia coli (Ec), and Staphylococcus epidermidis (Se), Pseudomonas aeruginosa (Pa) and the fungi Candida albicans (Ca). The identification results of the active association fungi were Circinelloides sp., Xylariaceae sp., Trichoderma asperellum, Aspergillus sp. and Pleosporales sp. . The extracts of these fungi had antioxidant activity with an IC50 value of 1071,62; 643,35; 1020,32; 805,70 and 784,31 mg/L. Furthermore, they also had an LC50 value of 462,67; 355,47; 504,92; 482,15 and 435,88 mg/L.
Keywords: Antimicrobial, Antioxidant, Fungi, Sponge, Symbiosis
Funding: The authors are grateful for the (DRPM) for providing the opportunity to conduct research.
Citation: Zainuddin, M., Pringgenies, D., Radjasa, O.K., and Haeruddin. 2022. Natural bioactive compounds of sponge-associated fungi with three marine ecosystems in Karimunjawa Island, Indonesia. CMU J. Nat. Sci. 21(1): e2022009.
INTRODUCTION
Control of infectious diseases has been carried out through the application of antibiotics, which are compounds that have the effect of suppressing or stopping a biochemical process in the organism. However, uncontrolled use of antibiotics can lead to the emergence of resistance and horizontal transfer of resistance to other pathogens through gene transfer (Von Wintersdorff et al., 2016; Lerminiaux and Cameron, 2019). There must be an effort to get new compounds that have the potential as antibacterial. On the other hand, it is known that the source of marine biological materials in Indonesia is abundant due to its geographical location and specific ecosystem so that the Indonesian marine area has very promising prospects for the future as a source of health pharmaceutical ingredients, which eventually leads to the pharmaceutical industry (Pringgenies et al., 2019).
There are many marine ecosystem specifications in Indonesia, namely coral reefs, seagrass (Unsworth et al., 2018), seaweed (Pringgenies et al., 2020) and mangrove ecosystems (Ariyanto et al., 2020). Each of these ecosystems has a high level of biodiversity because all three have different environmental characteristics (Ariyanto et al., 2019a), secondary metabolite (Ariyanto et al, 2019b; Ningsih et al., 2020) and marine organisms (Ariyanto et al., 2018; Santosa et al., 2020). These three ecosystems have certain ecological functions, one of which is as a habitat for vertebrate and invertebrate organisms such as sponges. Sponges are marine biota that produce secondary metabolites with various groups of terpenoids, acetogenins, alkaloids, cyclic halides, cyclic peptides, nitrogen compounds that are bioactive and have prospects in the field of pharmacology, such as their potential as antibacterial (Carroll et al., 2009). Secondary metabolites are not only produced by sponges, but also by their associated organisms. Sponges have associated microorganisms that provide food or produce certain metabolites that are beneficial to the sponge, which enter and are trapped in the pores (Sabdono and Radjasa, 2008).
Fungi are generally eukaryotic organisms composed of hyphae. Hyphae are divided into two types, namely septate hyphae and coenocytic hyphae. Septate hyphae have walls called septa (Steinberg et al., 2017). The septum divides the hyphae into cell-like units that are uninucleate (one nucleus). Meanwhile, coenocytic hyphae do not have septa and their nuclei mingle with each other. A group of hyphae forms a mycelium. This mycelium can then grow in different colors because the pigments produced by each Fungi are different.
The characteristics of the fungi that have been obtained can be seen macroscopically and microscopically (Hyde et al., 2000). The macroscopic characteristics are such as mycelia, color, texture, margin, height and aerial hyphae characteristics. While the microscopic characteristics are the color, texture and formation of conidia. Many bioactive compounds produced by sponges are known as secondary metabolites and the association of marine microorganisms has contributed to most of the bioactive compounds because they can produce the same metabolites as their host (Proksch et al., 2002). The research conducted at this time is preliminary research, further research is still ongoing to obtain compounds that act as antifungals in the research samples. Based on this, this study aimed to obtain sponge-associated fungi from coral, seagrass and mangrove ecosystems. Furthermore, this study aimed to determine its potential as an MDR antibacterial and identify the sponge-associated fungi which has potential as an antibacterial Multi Drug Resistant (MDR) by molecular methods.
MATERIALS AND METHODS
Study Design
The research was conducted on March – September 2020. The materials used in this study were sponges and microbial associations of sponges obtained from the coast of the Karimunjawa islands, Jepara Regency. Sampling was carried out at three ecosystem locations, namely coral reef ecosystems, seagrass ecosystems and mangrove ecosystems. The isolates used for the antibacterial activity test were isolates of MDR Staphylococcus aureus (Sa), Escherichia coli (Ec), and Staphylococcus epidermidis (Se), Pseudomonas aeruginosa (Pa) and the fungi Candida albicans (Ca).
Roadmap and Research Flowchart
Research on the study of the ecology of sponge-associated fungi and their potential as candidates for producing antibacterial compounds, with the following description: morphological collection and identification. Sponge samples were brought to the laboratory for isolation and purification of association fungi. The isolates obtained were subjected to a qualitative MDR antibacterial test. Furthermore, the results of the isolation of the sponge fungi were observed for morphological characteristics. The isolates with the best extract activity were identified molecularly.
Research Flow
The research was conducted in 6 stages, namely sampling, microbial isolation and purification, culture and extraction, bioassay (antibacterial, antifungal, antioxidant and BSLT toxicity tests), molecular identification and TLC and phytochemicals.
Identification of Sample
Sponge Identification
Identification was using the Hooper method (Hooper and Soest, 2002). Observation of external shape, color, oscula, spicules and surface. morphological form verified on sponge identification portal http://spongeguide.org and http://www.marinespecies.org/porifera/porifera.php?p= specimens.
Sponge-fungi isolation
Sponge symbiont fungi collection was done by cleaning the surface of the sponge by spraying it with sterile seawater. Then the sponge sample was cut into small pieces using a sterile cutter and then planted into a petri dish containing MEA solid medium aseptically then incubated for 2 x 24 h to get the sponge symbiont mushroom culture (Radjasa et al., 2007).
Fungi Morphological Characterization
Using the method of (Barnett and Hunter, 1998). Morphological characteristics included macroscopic observations: colony color, colony turning color, radial stripes, texture (granules, like flour, pile up and smooth). Microscopic observation: presence or absence of septa on hyphae, and characteristics of spores/conidia.
Cultivation of vegetative culture
A total of 2 loops of association fungi isolate were inoculated into 100 mL of PDY media in 250 mL Erlenmeyer. Culture with a temperature condition of 28 °C, agitation 150 rpm, for 2 days.
Antifungal activity test
Sponge-associated fungi isolate samples were extracted using the agar diffusion method. The agar medium used was PDA. The extract was dripped on a paper disk and placed on the surface of the media that had been inoculated with pathogenic fungi. The antifungal activity was carried out using the agar diffusion method. The agar medium used was Potato Dextrose Agar (PDA), the extract was dripped on a paper disk and placed on the surface of the media that had been inoculated with pathogenic fungi. The antifungal activity was indicated by the formation of a clear zone around the paper disk. The clear zone formed was then measured in diameter.
Molecular identification of isolates of each genomic cluster
Using the Radjasa method (Radjasa et al., 2007). The primer used was 18F (5’-ATC TGG TTG ATC CTG CCA GT-3’), and 18R primer (5’-GAT CCT TCC GCA GGT TCA CC-3’). The steps consisted of DNA preparation using Kappa ready mix master kit for PCR with amplification and evaluation by UV observation electrophoresis. Purification and sequencing of PCR products were carried out by PT. Genetic Science Indonesia using the same primer. Determination of species or genus, nucleotide sequences of each sample were analyzed using the BioEdit Sequence Alignment Editor version 7.0.9.1. Sequences were aligned using the Clustal W Multiple Alignment program and compared with MycoBank DNA data (http://www.mycobank.org) and BLAST data (http://www.blast.ncbi.nlm.nih.gov/blast). The aligned data was then reconstructed using maximum-parsimony analysis to obtain a phylogenetic tree using the Mega5 program with bootstrap values calculated from 1000 replications (Tamura et al., 2007).
Brine Shrimp Lethality Test (BSLT)
Toxicity was using the BSLT method (Costa-Lotufo et al., 2005). Concentration extracts of 100, 50 and 25 mg/L were pipetted as much as 50 μL into a vial, incubated, evaporated and added 20 μl of DMSO. A total of 10 samples / 5 mL of A. salina were put into a vial, then were incubated under an 18 W TL lamp for 24 h. Observation of the number of live and dead A. salina larvae. Percentage of deaths was calculated using the formula by (Meyer and Kuever, 2008).
Thin layer chromatography (TLC)
The extract solution was filed at the starting point on the TLC plate Aluminum Sheets Gel 60 F254 produced by Merck. Then the elution was plated with the solvent in the chromatography box (chamber) so that the compound moved to the maximum limit line. Observation of the spot was under the UV light of 256 and 365 nm. The retardation factor (Rf) value of each formed spot was calculation.
Antioxidant analysis of fungal extracts using the DPPH method
The DPPH free radical scavenging test was carried out with reference to (Leong and Shui, 2002) The percentage of DPPH free radical scavenging was calculated using the formula by (Rahman et al., 2015).
RESULTS
Characteristics of sponge-associated fungi
The results of isolation and purification of sponge association fungi from three ecosystems found 80 isolates of pure fungi, which were: 20 isolates from coral sponge-associated fungi, 34 isolates from seagrass-associated fungi and 26 isolates from mangrove-associated fungi. The results showed that the isolation of sponge-associated fungi was successfully isolated from 7 sponge samples in coral reef ecosystems. Samples of sponge type Callyspongia aerizusa had 5 isolates with the codes: K.S1.F1; K.S1.F2; K.S1.F3; K.S1.F4; and K.S1.F5. Sponge sample of Clathria reinwardti had 1 isolate, with the code K.S2.F1. The sample of sponge type Tectitethya sp. had 3 isolates coded K.S3.F1; K.S3.F2 and K.S3.F3. The sample of sponge type Halisarca sp. had 2 isolates coded K.S4.F1 and K.S4.F2. The sponge sample of Dercitus sp. type had 4 isolates, with the code K.S5.F1; K.S5.F2; K.S5.F3 and K.S5.F4. The sponge sample of Clathria sp. had 2 isolates coded K.S6.F1 and K.S6.F2. Samples of sponge species Speciospongia inconstans had 3 isolates coded K.S7.F1; K.S7.F2 and K.S7.F3.
Sponge samples in seagrass ecosystems obtained 11 isolates of sponge fungi, namely the sample of sponge type Agelas conifera which had 2 codes: L.S1.F1 and L.S1.F2. The sample of the Plakinastrella onkodes sponge had 4 isolates coded: L.S3.F1; L.S3.F2; L.S3.F3 and L.S3.F4. Samples of sponge species Chalinula pseudomolitba had 4 isolates coded L.S4.F1; L.S4.F2; L.S4.F3 and L.S4.F4. The sample of sponge species Clathria reinwardti had 5 isolates coded: L.S5.F1; L.S5.F2; L.S5.F3; L.S5.F4 and L.S5.F5. The sample of sponge type Tectitethya crypta had 5 isolates coded: L.S6.F1; L.S6.F2; L.S6.F3; L.S6.F4 and L.S6.F5. Samples of sponge type Amphimedon viridis had 4 isolates coded: L.S8.F1; L.S8.F2; L.S8.F3 and L.S8.F4. The sample of the sponge type Haliclone cymaeformis had 3 isolates coded L.S9.F1; L.S9.F2 and L.S9.F3. Sample of sponge type Halisarca sp. had 5 isolates coded L.S10.F1; L.S10.F2; L.S10.F3; L.S10.F4 and L.S10.F5. While sponge sample of Hyrtios violaceus had 2 isolates coded L.S11.F1 and L.S11.F2.
There were 9 sponge samples from the mangrove ecosystem, namely the C. pseudomolitba sponge sample which had 5 isolates coded; M.S2.F1; M.S2.F2; M.S2.F3; M.S2.F4 and M.S2.F5. Sample of sponge type T. crypta had 4 isolates coded M.S4.F1; M.S4.F2; M.S4.F3 and M.S4.F4. The sponge sample type H. melanododa had 5 isolates coded M.S5.F1; M.S5.F2; M.S5.F3; M.S5.F4 and M.S5.F5. Samples of sponge type A. viridis have 2 isolates coded M.S6.F1 and M.S6.F2. Sample of the sponge type S. vesparium had 5 isolates coded M.S7.F1; M.S7.F2; M.S7.F3; M.S7.F4 and M.S7.F5. While the sponge sample type Haliclona oculata had 5 isolates coded M.S9.F1; M.S9.F2; M.S9.F3; M.S9.F4 and M.S9.F5.
The next step was to culture sponge-associated fungi into Potato Dextrose Broth (PDB) liquid medium. Fungi were seen forming biomass on the surface of the medium. Fungal mycelia covered the entire surface of the liquid medium. At the end of the incubation period, the fungal mycelia tissue was taken and separated with the liquid medium. The fungal mycelia biomass obtained was prepared and macerated using methanol solvent. A total of 80 isolate extracts of sponge-associated fungi were tested against MDR test bacteria, which were S.s aureus, S. epidermidis, E. coli, P. aeruginosa and one type of fungal pathogen, C.albicans.
The antifungal activity test of fungal isolates was carried out using the agar diffusion method, the results showed that the sponge association fungi isolate had antifungal activity. The antibacterial activity test of the isolates against the pathogenic S. aureus showed 6 isolates that were active against the pathogenic bacteria, which were 5 isolates from the seagrass ecosystem: L.S3.F4; L.S5.F4; L.S6.F2; L.S8.F2; L.S11.F2 and 1 isolate from the mangrove ecosystem M.S4.F2. L.S11.F2 isolate had the highest inhibition zone, which was 1.77 mm against S. aureus. The antibacterial activity test of fungal isolates against the pathogenic bacteria S. epidermidis showed that there were 6 active isolates, which were 2 isolates of fungi from coral reef ecosystems: K.S3.F2; K.S6.F2, 2 isolates from seagrass ecosystem: L.S3.F1; L.S8.F2 and 2 isolates from mangrove ecosystem: M.S2.F4, M.S9.F2. The isolate M.S9.F2 had the highest inhibition zone against the test bacteria S. epidermidis, which was 1.45 mm. Antibacterial activity test of sponge-associated fungi isolates against pathogenic bacteria E. coli showed 4 active isolates, which were 2 isolates from coral ecosystem: K.S6.F2, K.S7.F1, 1 isolate from seagrass ecosystem: L.S8.F2, and 1 isolate from mangrove ecosystem: M.S2.F2. K.S7.F1 isolate had the highest zone of inhibition against the test bacteria E.coli: 1.54 mm. The antibacterial activity test of fungal isolates against pathogenic bacteria P. aeruginosa showed 2 active isolates, namely 1 isolate from seagrass ecosystems: L.S8.F2 which had a higher inhibition zone (1.39 mm) than an isolate from mangrove ecosystems: M.S7.F5 (1.32mm).
The antifungal activity test of sponge-associated fungi isolates against the pathogenic fungi C. albicans showed that there were 5 active isolates, namely 1 isolate from the coral ecosystem: K.S1.F1; L.S8.F2, 2 isolates from seagrass ecosystem: K.S1.F1; L.S8.F2, and 2 isolates from mangrove ecosystem: M.S2.F4 and M.S2.F4. The isolate M.S2.F4 had the highest inhibition zone against the test fungus C. albicans, which was 1.67 mm. The activity test of the sponge-associated fungi isolates against the test bacteria and the test fungi can be seen in the Table 1.
Table 1. Results of the activity of the sponge-associated fungi isolates against bacteria and fungi.
|
No |
Isolates |
Multi Drug Resistant (mm) |
||||
|
S. aureus |
S.epidermidis |
E.coli |
P.aeruginosa |
C.albicans |
||
|
1 |
K.S1.F1 |
- |
- |
- |
- |
1,37 |
|
2 |
K.S3.F2 |
- |
1.38 |
- |
- |
- |
|
3 |
K.S6.F2 |
- |
1.41 |
1.37 |
- |
- |
|
4 |
K.S7.F1** |
- |
- |
1.54* |
- |
- |
|
5 |
L.S3.F1 |
- |
1.43 |
- |
- |
- |
|
6 |
L.S3.F4 |
1.40 |
- |
- |
- |
- |
|
7 |
L.S5.F4 |
1.36 |
- |
- |
- |
- |
|
8 |
L.S6.F2 |
1.34 |
- |
- |
- |
- |
|
9 |
L.S8.F2** |
1.64 |
1.39 |
1.41 |
1.39* |
1.51 |
|
10 |
L.S10.F4 |
- |
- |
- |
- |
1.28 |
|
11 |
L.S11.F2** |
1.77* |
- |
- |
- |
- |
|
12 |
M.S2.F1 |
- |
- |
- |
- |
1.34 |
|
13 |
M.S2.F2 |
- |
- |
1.47 |
- |
- |
|
14 |
M.S2.F4** |
- |
1.33 |
- |
- |
1.67* |
|
15 |
M.S4.F2 |
1.35 |
- |
- |
- |
- |
|
16 |
M.S7.F5 |
- |
- |
- |
1.32 |
- |
|
17 |
M.S9.F2** |
- |
1.45* |
- |
- |
- |
Based on the results of the study in Table. 1, there were 17 isolates of sponge-associated fungi that had activity. For more details, 17 fungi active isolates against pathogenic bacteria and fungal pathogens were described in terms of morphological characteristics of sponge-associated fungi as shown in Table 2.
Table 2. Morphological characteristics of sponge-associated fungi.
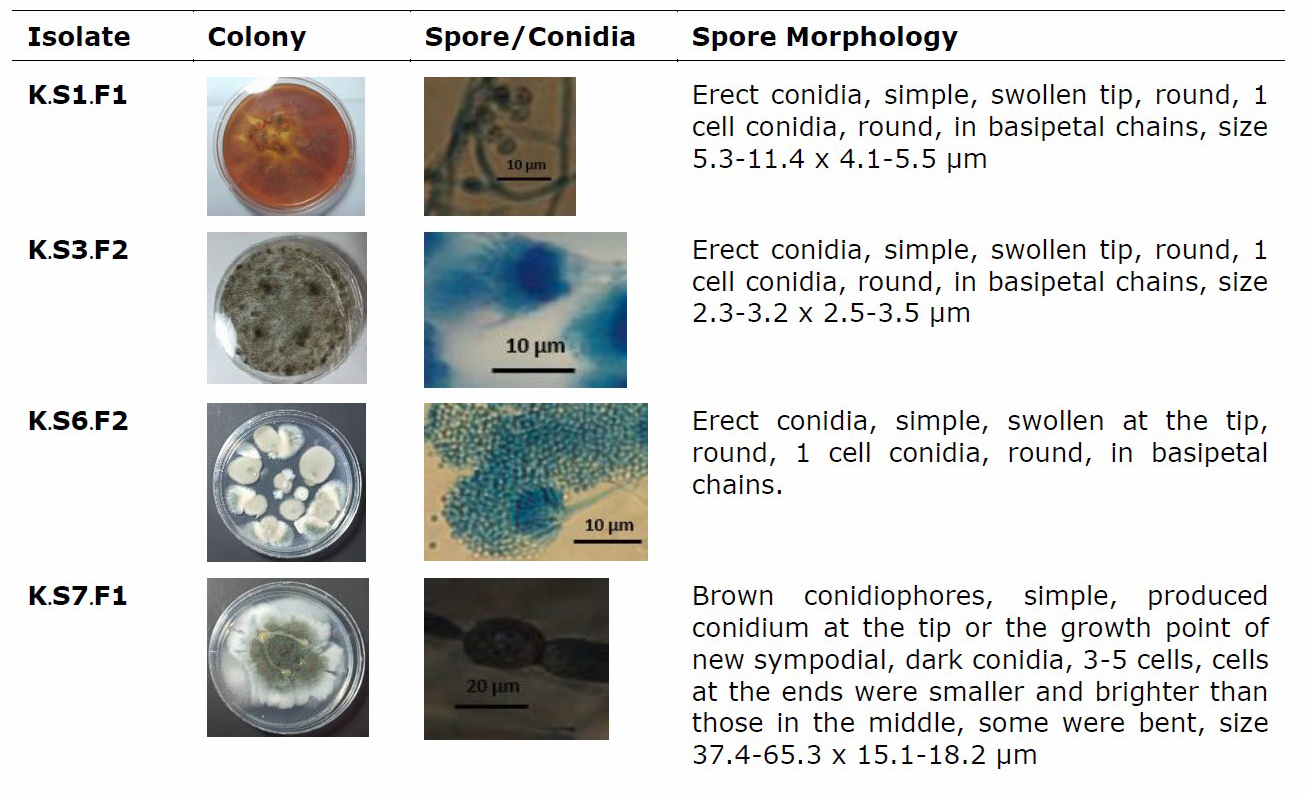
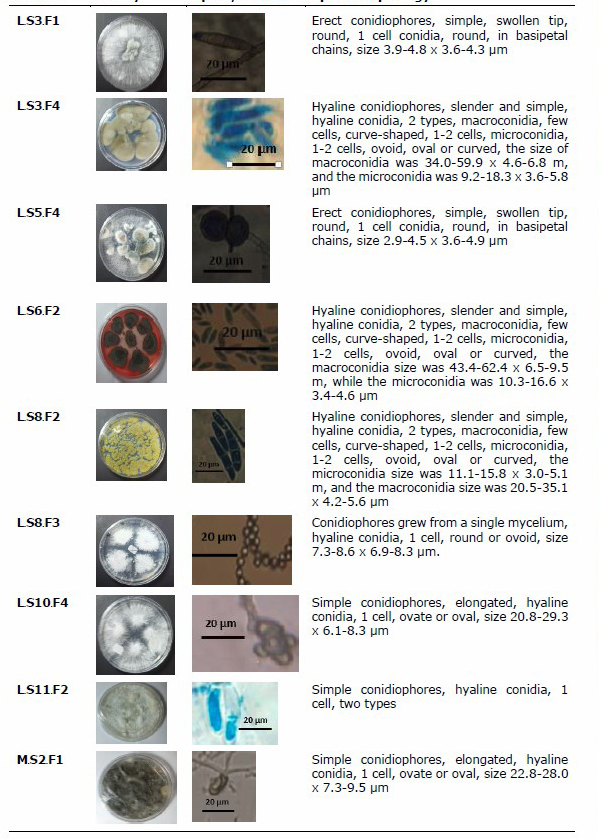

The 17 isolates of sponge-associated fungi, 5 isolates of associated fungi that had the best zone values were selected, namely isolates K.S7.F1; L.S8.F2; L.S11.F2; M.S2.F4 and M.S9.F2, and then were identified by molecular methods. Based on the results of the study, it was shown that the PCR process was successfully carried out with good gel doc visualization results. The PCR results showed a single and uncontaminated band and then a sequencing analysis was performed to obtain the acid and base sequences of fungal isolate DNA for homology BLAST. The BLAST results are presented in Table 3.
Table 3. Results of BLAST homology identification of sponge-associated fungi.
|
No. |
Isolate |
Sequence (bp) |
Bacteria Species Name |
Homology (%) |
No. Access |
|
1. |
K.S3.F2 |
627 |
Circinelloides strain OUCMBI101096 |
99 |
HQ914900.1 |
|
2. |
L.S8.F2 |
582 |
Xylariaceae sp. B1a0413SNA2CC912 |
97 |
KP322785.1 |
|
3. |
L.S11.F2 |
255 |
Trichoderma asperellum |
99 |
MG669099.1 |
|
4. |
M.S2.F4 |
396 |
Aspergillus sp. |
98 |
MK775137.1 |
|
5. |
M.S9.F2 |
256 |
Pleosporales sp. B1A0415P152CC716 |
99 |
KP322784.1 |
Based on the results of the study, it showed that 5 isolates had a high percentage of homology, which was around 97-99%. The isolate with a homology value of 97% was L.S8.F2, the isolate had a sequence value of 582 which was identified as Xylariaceae sp. B1a0413SNA2CC912 with access number KP322785.1. The isolate with a homology value of 98% was M.S2.F4, the isolate had a sequence value of 396 which was identified as B Aspergillus sp. with access number MK775137.1. Isolates with 99 % homology value were isolates K.S3.F2, L.S11.F2 and M.S9.F2. The K.S3.F2 isolate had a sequence value of 627 bp with access no HQ914900.1, identified as Circinelloides strain OUCMBI101096. Isolate L.S11.F2 had a sequence value of 255 bp, with the access number MG669099.1 and was identified as T. asperellum. Isolate M.S9.F2 had a sequence value of 256 bp with the access number KP322784.1 and was identified as Pleosporales sp. B1A0415P152CC716.
Furthermore, 5 isolates of the selected association fungi were tested for cytotoxicity of the extract against the organism Artemia salina using the Brine Shrimp Lethality Test (BSLT) method. The results of the cytotoxicity test using the BSLT method were presented in Table 4.
Table 4. Cytotoxicity analysis of fungal extracts using the BSLT method.
|
No |
Isolate |
Concentration (mg/L) |
Regression |
R2 |
LC50 (mg/L) |
|||||
|
0 |
10 |
100 |
250 |
500 |
750 |
|||||
|
1 |
K.S3.F2 |
5 |
12 |
17 |
40 |
52 |
73 |
y = 0,0864x + 10,025 |
0,973 |
462,67 |
|
2 |
L.S8.F2 |
7 |
10 |
20 |
31 |
74 |
94 |
y = 0,1192x + 7,6278 |
0,988 |
355,47 |
|
3 |
L.S11.F2 |
5 |
11 |
18 |
32 |
43 |
73 |
y = 0,0827x + 8,2433 |
0,973 |
504,92 |
|
4 |
M.S2.F4 |
6 |
13 |
16 |
37 |
49 |
72 |
y = 0,0831x + 9,9333 |
0,978 |
482,15 |
|
5 |
M.S9.F2 |
8 |
12 |
18 |
35 |
52 |
81 |
y = 0,0932x + 9,3760 |
0,991 |
435,88 |
Results of the cytotoxicity test using the BSLT method showed that the fungal extract of K.S3.F2 isolate at concentrations of 0, 10, 100, 250, 500 and 750 mg/L had toxicity values of 5%, 12%, 17%, 40%, 52% and 73 %. The extract toxicity pattern formed a linear regression with the equation y = 0.0864x + 10.025 with a determinant coefficient of 0.973. Based on this equation, the value of the LC50 extract was 462.67 mg/L. Fungal extract of isolate L.S8.F2 with concentrations of 0, 10, 100, 250, 500 and 750 ppm had the toxicity values at 7%, 10%, 20%, 32%, 74% and 94%. The extract toxicity pattern formed a linear regression with the equation y = 0.1192x + 7.6278 and a determinant coefficient of 0.988. Based on this equation, the value of the LC50 extract was 355.47 mg/L. Fungal extract of isolate L.S11.F2 with the concentrations of 0, 10, 100, 250, 500 and 750 ppm had the toxicity values at 5%, 11%, 18%, 32%, 43% and 73%. The extract toxicity pattern formed a linear regression with the equation y = 0.0827x + 8.2433 and a determinant coefficient of 0.973. Based on this equation, the value of the LC50 extract was 504.92 mg/L. Fungal extract of isolate M.S2.F4 at concentrations of 0, 10, 100, 250, 500 and 750 ppm had the toxicity values at 6%, 13%, 16%, 37%, 49% and 72%. The extract toxicity pattern formed a linear regression with the equation y = 0.0831x + 9.9333 and a determinant coefficient of 0.978. Based on this equation, the value of the LC50 extract was 482.15 mg/L. Isolate M.S9.F2 fungi extract at concentrations of 0, 10, 100, 250, 500 and 750 mg/L had the toxicity values at 8%, 12%, 18%, 35%, 52% and 81%. The extract toxicity pattern formed a linear regression with the equation y = 0,0932x + 9.3760 and a determinant coefficient of 0,991. Based on this equation, the value of LC50 extract was 435,88 mg/L.
Research had been carried out and found the best 5 extracts in their activity as antifungals. Furthermore, the five extracts were extracts from the association fungi isolate K.S7.F1; L.S8.F2; L.S11.F2; M.S2.F4 and M.S9.F2, which were tested for antioxidant activity using the DPPH method. The results of the antioxidant test using the DPPH method are presented in Table 5.
Table 5. Antioxidant analysis of fungal extracts using the DPPH method.
|
No |
Isolate |
Concentration (mg/L) |
Regression |
R2 |
IC50 (mg/L) |
|
|||||
|
100 |
250 |
500 |
1000 |
2500 |
|
||||||
|
1 |
K.S3.F2 |
29 |
40 |
40 |
51 |
73 |
y = 0,0167x + 32,104 |
0,965 |
1071,62 |
||
|
2 |
L.S8.F2 |
30 |
40 |
50 |
63 |
95 |
y = 0,0254x + 33,659 |
0,972 |
643,35 |
||
|
3 |
L.S11.F2 |
20 |
31 |
41 |
53 |
85 |
y = 0,0251x + 24,390 |
0,970 |
1020,32 |
||
|
4 |
M.S2.F4 |
26 |
37 |
47 |
58 |
90 |
y = 0,0249x + 29,938 |
0,972 |
805,70 |
||
|
5 |
M.S9.F2 |
31 |
41 |
49 |
55 |
83 |
y = 0,0195x + 34,706 |
0,962 |
784,31 |
||
The results showed that the fungal extract of K.S3.F2 isolate at concentrations of 100, 250, 500, 1000 and 2500 ppm had radical reduction values of 29%, 40%, 40%, 51% and 73%. The extract inhibition pattern formed a linear regression with the equation y = 0,0167x + 32,104 with a determinant coefficient of 0,965. Based on this equation, the extract IC50 value was 1071,62 mg/L. L.S8.F2 isolate fungi extract at concentrations of 100, 250, 500, 1000 and 2500 ppm had radical reduction values of 30%, 40%, 50%, 63% and 95%. The extract inhibition pattern formed a linear regression with the equation y = 0,0254x + 33,659 with a determinant coefficient value of 0,972, so that the equation obtained the extract IC50 value of 643,35 ppm. Fungal extracts isolates L.S11.F2 at concentrations of 100, 250, 500, 1000 and 2500 mg/L had radical reduction values of 20%, 31%, 41%, 53% and 85%. The extract inhibition pattern formed a linear regression with the equation y = 0,0251x + 24,390 with a determinant coefficient of 0,970. Based on this equation, the extract IC50 value was 1020,32 mg/L. M.S2.F4 isolate fungi extract at concentrations of 100, 250, 500, 1000 and 2500 mg/L had radical reduction values of 26%, 37%, 47%, 58% and 90%. The extract inhibition pattern formed a linear regression with the equation y = 0,0249x + 29,938 with a determinant coefficient of 0,972. Based on this equation, the extract IC50 value was 805,70 mg/L. M.S9.F2 isolate fungi extract at concentrations of 100, 250, 500, 1000 and 2500 mg/L had radical reduction values of 31%, 41%, 49%, 55% and 83%. The extract inhibition pattern formed a linear regression with the equation y = 0.0195x + 34.706 with a determinant coefficient of 0,962. Based on this equation, the extract IC50 value was 784,31 mg/L.
Results of the toxicity test and antioxidant activity test showed that the extract with the best toxicity value and antioxidant activity was the extract from the association fungi isolate L.S8.F2. The extract from association fungi L.S8.F2 had an LC50 value of 355,47 mg/L and an IC50 value of 643,35 mg/L. Then, the study conducted a culture of association fungi L.S8.F2 to obtain mycelial biomass and post-culture medium for extraction. Mycelia extract and association fungal culture medium extract L.S8.F2 were tested by Thin Layer Chromatography (TLC) to determine the pattern and composition of the compound spots in it. Results of the TLC test activity showed that the extract of the mycelia fungi association isolate L.S8.F2 had 5 single spots, which were spot 1 with yellow-orange color with an Rf value of 0,08; spot 2 which was green and yellow with an Rf value of 0,24; spot 3 which was light green with an Rf value of 0,31; spot 4 which was dark green with an Rf value of 0,37; and spot 5 which was green-blue with an Rf value of 0,54 as shown in Figure 1.
The extract from the post-culture medium of the L.S8.F2 isolate had 6 single spots, namely spot 1 which was yellow with an Rf value of 0,07; spot 2 which was light blue with an Rf value of 0,16; spot 3 which was blue-green with an Rf value of 0,25; spot 4 which was light green with an Rf value of 0,36; spot 5 which was dark green with an Rf value of 0,61; and spot 6 which was dark green with an Rf value of 0,72.
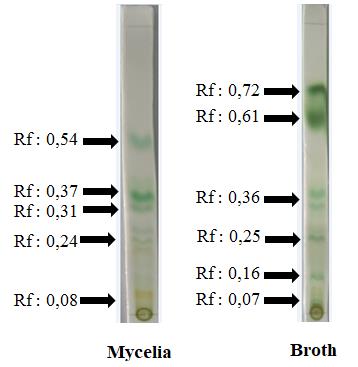
Figure 1. TLC analysis of mycelia extracts and culture broth of L.S8.F2 fungal isolates.
Based on the results of the phytochemical analysis of mycelia extract and culture broth of the L.S8.F2 isolate, it was shown that the extract with the best toxicity value and antioxidant activity was the extract from the association fungi isolate L.S8.F2. The extract of the association fungi L.S8.F2 was tested by TLC, then the mycelia extract and the post-culture medium were subjected to phytochemical analysis. Phytochemical analysis was carried out qualitatively with several reagents. Phytochemical analysis carried out included testing for the presence of alkaloids, phenols, flavonoids, saponins and steroids. Phytochemical test of alkaloid compounds was carried out using 3 methods, which were the Dragendorf, Mayer and Wagner methods. Results of the phytochemical test of mycelia extract and post-culture medium from the association fungi L.S8.F2 were presented in Table 5. Based on the results of the study, it was shown that the mycelia extract and post-culture medium from the association fungi L.S8.F2 contained several groups of phytochemical compounds. Mycelia extract from association fungi isolate L.S8.F2 contained phenol and flavonoid group compounds. Meanwhile, the post-culture medium extract from association fungi isolate L.S8.F2 contained alkaloids, phenols and flavonoids.
DISCUSSION
The research carried out was to isolate fungi associated with sponges in coral, seagrass and mangrove ecosystems. The isolation results found 80 isolates of pure fungi, namely: 20 isolates from coral sponge-associated fungi, 34 isolates from seagrass association fungi and 26 isolates from mangrove association fungi. Results of the study found that sponges had symbiotic fungi and were successfully isolated in the laboratory. The symbiotic fungi in sponges had a symbiotic relationship and functioned ecologically. The form of this symbiotic relationship was the relationship between the host organism that provides organic nutrients, a place to live and protect the symbiotic fungi. The form of the symbiotic relationship was specific. Symbiont fungi produce metabolic compounds so that they can adapt to their host organisms (Bhagobaty & Joshi, 2012). This was also stated in the results of research by (Elbandy et al., 2009) who succeeded in isolating the symbiont fungus P. illacinus which ecologically has an association relationship with the sponge Petrosia sp. Besides, there were also fungi Paecilomyces sp., Fusarium sp. and Penicillium spp. which can be isolated from the Tethya aurantium sponge (Wiese et al., 2011). Edrada et al., (2002) isolated and purified the fungus P. ontanense from the symbiotic X. Exigua sponge in marine waters. The fungus H. werneckii which is symbiotic with the sponge Aplysina aerophoba had also been successfully isolated on a laboratory scale (Brauers et al., 2001). Liu et al., (2010) succeeded in isolating the symbiotic fungus P. shrysogenum from the sponge Gelliodes carnosa, while it had obtained isolates of the symbiont fungus A. versicolor from the sponge Haliclona simulans. In addition to sponges, several types of symbiotic fungi were also found, including Penicillium spp. isolated from Green algae C. racemosea and C. scalpelliformisa. Fungi Fusarium spp. was isolated from green seaweeds C. racemosaa and C. sertularioides (Suryanarayanan, 2012).
Results of the research that had been carried out showed that the symbiont fungi extracts K.S7.F1; L.S8.F2; L.S11.F2; M.S2.F4 and M.S9.F2 had antimicrobial activity against Staphylococcus aureus, S. epidermidis, Escherichia coli, P. aeruginosa and one type of fungal pathogen, C. albicans. This was presumably because the symbiont fungi extract contains bioactive compounds that are bactericidal and fungicidal. The same thing was found in several studies. Fungal extract P. oxalicum at a concentration of 100 µg/disk has a growth inhibition activity against pathogenic bacteria E. coli by 19 mm (Suryanarayanan, 2012). The study of extracts at the concentration of 100 µg/disk showed inhibitory activity against P.solanacearum, X. campestris, A. tumefaciens, E. coli, S. marcescens (Paul et al., 2012).
Several studies of fungal extracts of P. oxalicum had antifungal activity against the fungal pathogen A. sydowii with an inhibition zone of 12 mm. Besides, it also inhibited the growth of the fungi C. acutatum, F. oxysporum, Phytophthora sapsici (Paul et al., 2012). Some researchers had succeesed in isolating the fungus P. oxalicum from sponges and extracting fungi symbionts using the organic solvent ethyl acetate. The results of the identification of compounds had found xanthan, penioxalicin and chromone compounds that have antibacterial activity (Bao et al., 2014; Bian et al., 2015). Fungal extract Penicillium sp. which was in symbiosis with the brown seaweed X. gladiate contained the alkaloid compound 2-pyridone and provided antimicrobial activity at a concentration of 30 g/disk against the bacteria B. subtilis and the fungus C. albicans by 8 mm and 11 mm, respectively. Fungal extract Penicillium sp. also had a toxicity with IC50 value of 1.8 g/mL against leukemia test activity (Da Silva et al., 2006).
Aspergillus niger fungi extract in symbiosis with the tunicate Aplindium sp. had antibacterial activity sensitive to S. aureus and E. faecium. Aspergillus niger fungi extract contained compounds yanuthones, epoxycylohexenones, 1-hydroxy-yanuthone, 22-deacetyl-yanuthone and 1-hydroxy-yanuthone (Bugni and Ireland, 2004). Results of the fungal symbionts identification in the study showed that the isolates were Circinelloides, Xylariaceae sp., T. asperellum, Aspergillus sp. and Pleosporales sp.. The circinelloides fungus is one type of mucor that can be isolated from clinical sources. In addition, mucor is used in the field of food production biotechnology. The circinelloides fungi have a filamentous morphology, and under certain conditions have growth in the form of yeast. Xylariaceae sp. is a symbiont containing antimicrobial compounds. Xylariaceae sp. also has inhibitory activity against enzymes.
T. asperellum is a symbiotic fungus that has antibacterial activity by overlay method against E. coli pathogens. Methanol extract from the mycelium of the fungus Trichoderma asperellum had antibacterial activity and contained phytochemical compounds from the hydroquinone and flavonoid groups of phenolic compounds. Ethyl acetate extract from the broth of the T. asperellum fungal culture process contained phytochemical compounds from the hydroquinone group of phenols, saponins, alkaloids and flavonoids. The content of saponins, flavonoids are also found in many plants, such as those found in fruits of Emblica, Yellow Myrobalan and Bastard Myrobalan (Thamapan et al., 2020).
Trichoderma originating from the sea grew slowly because of the difficulty of isolating the fungus. In our study, 30 strains of marine-derived Trichoderma were identified through the translational sequence of elongation factor 1-alpha (EF1_A), and their biological activities, such as antioxidant activity by ABTS and DPPH assays, antifungal activity against Asteromyces cruciatus and Lindra thalassiae, also tyrosinase inhibitory activity were investigated. T. asperellum fungal extract had antioxidant activity by ABTS method and also had antifungal activity. Fungal extracts of T. bissettii and T. guizhouense had anti-oxidant activity by reducing DPPH free radicals and inhibiting tyrosinase activity (Da Silva et al., 2006). Other information about the fruit contains antioxidants that provide potential treatment and prevention for diabetes with benefits on the innate defense system Cydonia oblonga (Sakhri et al., 2021). The interesting thing about the results of this study is that in addition to having anti-oxidant content, it also functions as an antifungal.
CONCLUSION
The research had succeeded in isolating the association fungi of Circinelloides, Xylariaceae sp., T. asperellum, Aspergillus sp. and Pleosporales sp.. The ethyl acetate extract of the fungus had antibacterial and antioxidant activity. Sponge-associated fungi extract had a class of phytochemical compounds that have potential prospective in the pharmaceutical field.
ACKNOWLEDGEMENT
We would like to thank the Ministry of Research, Technology and Higher Education (DRPM): NO SPK : 225-17/UN7.6.1/PP/2020 for providing the opportunity to conduct research.
AUTHOR CONTRIBUTIONS
Delianis Pringgenies, Ocky Karna Radjasa and Haeruddin assisted in conducting the experiments, performed the statistical analysis and data visualization and wrote the manuscript. Muhammad Zainuddin and Delianis Pringgenies designed and conducted all of the experiments and wrote the manuscript. All authors have read and approved of the final manuscript.
CONFLICT OF INTEREST
The authors declare that they hold no competing interests.
REFERENCES
Ariyanto, D., Bengen, D.G., Prartono, T., and Wardiatno, Y. 2018. The association of Cassidula nucleus (Gmelin 1791) and Cassidula angulifera (petit 1841) with mangrove in banggi coast, Central Java, Indonesia. AACL Bioflux. 11: 348–361.
Ariyanto, D., Bengen, D.G., Prartono, T., and Wardiatno, Y. 2019a. The physicochemical factors and litter dynamics (Rhizophora mucronata lam. and Rhizophora stylosa griff) of replanted Mangroves, Rembang, Central Java, Indonesia. Environment and Natural Resources Journal. 17: 11–29.
Ariyanto, D., Gunawan, H., Puspitasari, D., Ningsih, S.S., Jayanegara, A., and Hamim, H. 2019b. The differences of the elements content in Rhizophora mucronata leaves from Asahan Regency, North Sumatra, Indonesia. Polish Journal of Natural Sciences. 34: 481–491.
Ariyanto, D., Bengen, D.G., Prartono, T., and Wardiatno, Y. 2020. Distribution and abundance of Cerithideopsilla djadjariensis (Martin 1899) (potamididae) on Avicennia marina in Rembang, Central Java, Indonesia. Egyptian Journal of Aquatic Biology and Fisheries. 24: 323–332.
Bao, J., Luo, J.F., Qin, X.C., Xu, X.Y., Zhang, X.Y., Tu, Z.C., and Qi, S.H. 2014. Dihydrothiophene-condensed chromones from a marine-derived fungus Penicillium oxalicum and their structure-bioactivity relationship. Bioorganic and Medicinal Chemistry Letters. 24: 2433–2436.
Barnett, H.L., and Hunter, B.B. 1998. A comprehensive resource for recognizing, identifying, and learning various aspects of imperfect fungi.
Bhagobaty, R.K., and Joshi, S.R. 2012. Enzymatic activity of fungi endophytic on five medicinal plant species of the pristine sacred forests of meghalaya, India. Biotechnology and Bioprocess Engineering. 17: 33–40.
Bian, X., Bai, J., Hu, X., Wu, X., Xue, C., Han, A., Su, G., Hua, H., and Pei, Y. 2015. Penioxalicin, a novel 3-nor-2,3-seco-labdane type diterpene from the fungus Penicillium oxalicum TW01-1. Tetrahedron Letters. 56: 5013–5016.
Brauers, G., Ebel, R., Edrada, R.A., Wray, V., Berg, A., Gräfe, U., and Proksch, P. 2001. Hortein, a new natural product from the fungus Hortaea werneckii associated with the sponge Aplysina aerophoba. Journal of Natural Products. 64: 651–652.
Bugni, T.S., and Ireland, C.M. 2004. Marine-derived fungi: A chemically and biologically diverse group of microorganisms. Natural Product Reports. 21: 143–163.
Carroll, A.R., Duffy, S., and Avery, V.M. 2009. Citronamides A and B, tetrapeptides from the Australian sponge Citronia astra. Journal of Natural Products. 72: 764–768.
Costa-Lotufo, L.V., Khan, M.T.H., Ather, A., Wilke, D.V., Jimenez, P.C., Pessoa, C., De Moraes, M.E.A., and De Moraes, M.O. 2005. Studies of the anticancer potential of plants used in Bangladeshi folk medicine. Journal of Ethnopharmacology. 99: 21–30.
Da Silva, P.R.F., Argenta, G., Sangoi, L., Strieder, M.L., and Da Silva, A.A. 2006. Estratégias de manejo de coberturas de solo no inverno para cultivo do milho em sucessão no sistema semeadura direta. Ciencia Rural. 36: 1011–1020.
Edrada, R.A., Heubes, M., Brauers, G., Wray, V., Berg, A., Gräfe, U., Wohlfarth, M., Mühlbacher, J., Schaumann, K., Sudarsono, Bringmann, G., and Proksch, P. 2002. Online analysis of xestodecalactones A-C, novel bioactive metabolites from the fungus Penicillium cf. montanense and their subsequent isolation from the sponge Xestospongia exigua. Journal of Natural Products. 65: 1598–1604.
Elbandy, M., Shinde, P.B., Hong, J., Bae, K.S., Kim, M.A., Lee, S.M., and Jee H. Jun. 2009. a-Pyrones and Y 이 low Pigments from the Sponge-Derived Fungus Paecilomyces lilacinus. Bulletin of the Korean Chemical Society. 30: 188–192.
Hooper, J.N.A., and Soest, R.W.M. Van. 2002. Systema Porifera. Systema Porifera.
Hyde, W.T., Crowley, T.J., Baum, S.K., and Peltier, W.R. 2000. Neoproterozoic “snowball earth” simulations with a coupled climate/ice- sheet model. Nature. 405: 425–429.
Leong, L.P. and Shui, G. 2002. An investigation of antioxidant capacity of fruits in Singapore markets. Food Chemistry. 76: 69–75.
Lerminiaux, N.A. and Cameron, A.D.S. 2019. Horizontal transfer of antibiotic resistance genes in clinical environments. Canadian Journal of Microbiology. 65: 34–44.
Liu, W. C., Li, C. Q., Zhu, P., Yang, J.L., and Cheng, K.D. 2010. Phylogenetic diversity of culturable fungi associated with two marine sponges: Haliclona simulans and Gelliodes carnosa, collected from the Hainan Island coastal waters of the South China Sea. Fungal Diversity. 42: 1–15.
Meyer, B. and Kuever, J. 2008. Phylogenetic diversity and spatial distribution of the microbial community associated with the Caribbean deep-water sponge Polymastia cf. corticata by 16S rRNA, aprA, and amoA gene analysis. Microbial Ecology. 56: 306–321.
Ningsih S.S., Ariyanto, D., Puspitasari, D., Jayanegara A., Hamim, H., and Gunawan, H. 2020. The amino acid contents in mangrove Rhizophora mucronata leaves in Asahan, North Sumatra, Indonesia. E3S Web of Conferences. 151: 1-3
Paul, N.C., Deng, J.X., Sang, H.K., Choi, Y.P., and Yu, S.H. 2012. Distribution and antifungal activity of endophytic fungi in different growth stages of chili pepper (Capsicum annuum L.) in Korea. Plant Pathology Journal. 28:10–19.
Pringgenies, D., Retnowati, E.I., Ariyanto, D., Dewi, K., Viharyo, M.A.S., Susilowati, R. 2020. Symbiotic microbes from various seaweeds with antimicrobial and fermentative properties, Aquaculture, Aquarium, Conservation & Legislation. 2211-2217
Pringgenies, D., Yudiati, E., Djunaedi, A., Santosa, G.W., and Koesoemadji. 2019. Explorations of symbiotic microbe from sea cucumber gut as an anti-multi-drug resistant microbe agent for utilization in hand sanitizer products. AACL Bioflux. 12: 737–747.
Proksch, P., Edrada, R.A., and Ebel, R. 2002. Drugs from the seas – current status and microbiological implications. Applied Microbiology and Biotechnology. 59: 125–134.
Radjasa, O.K., Sabdono, A., Zocchi, J., and Zocchi, E. 2007. Richness of secondary metabolite-producing marine bacteria associated with sponge Haliclona sp. International Journal of Pharmacology. 3: 275–279.
Rahman, M.M., Islam, M.B., Biswas, M., and Khurshid Alam, A.H.M. 2015. In vitro antioxidant and free radical scavenging activity of different parts of Tabebuia pallida growing in Bangladesh. BMC Research Notes. 8: 1–9.
Sabdono, A. and Radjasa, O.K. 2008. Phylogenetic Diversity of Organophosphorous Pesticide-Degrading Coral Bacteria from Mid-West Coast of Indonesia. Biotechnology. 7: 694–701.
Santosa, G.W., Djunaedi, A., Susanto, A.B., Pringgenies, D., and Ariyanto, D. 2020. Characteristics of bioactive compounds of Holothuria atra (Jaeger, 1833) associated bacteria. AACL Bioflux. 13: 2161–2169.
Sakhri F.Z., Zerizer, S., and Bensouici, C. 2021. Evaluation of The Antioxidant, Antidiabetic and Immunomodulatory Activity of Cydonia oblonga Fruit Extract. Published: Apr 1, 2021 Chiang Mai University Journal of Natural Sciences. 20: e2021052.
Steinberg, G., Penalva, M.A., Riquelme, M., Wösten, H.A., and Harris, S.D. 2017. Cell biology of hyphal growth. The Fungal Kingdom. 231–265.
Suryanarayanan, T.S. 2012. The diversity and importance of fungi associated with marine sponges. Botanica Marina. 55: 553–564.
Tamura, K., Dudley, J., Nei, M., and Kumar, S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 24: 1596–1599.
Thamapan, K., Loahakunjit, N., Kerdchoechuen, O., Vongsawasdi, P., and Mingvanish, W., 2020. Phytochemical evaluation and qualitative analysis as sweetness potentially of selected thai plants. Chiang Mai University Journal of Natural Sciences. 19: 315-332.
Unsworth, R.K.F., Ambo-Rappe, R., Jones, B.L., La Nafie, Y.A., Irawan, A., Hernawan, U.E., Moore, A.M., and Cullen-Unsworth, L.C. 2018. Indonesia’s globally significant seagrass meadows are under widespread threat. Science of the Total Environment. 634: 279–286.
Von Wintersdorff, C.J.H., Penders, J., Van Niekerk, J.M., Mills, N.D., Majumder, S., Van Alphen, L.B., Savelkoul, P.H.M., and Wolffs, P.F.G. 2016. Dissemination of antimicrobial resistance in microbial ecosystems through horizontal gene transfer. Frontiers in Microbiology. 7: 1–10.
Wiese, J., Ohlendorf, B., Blümel, M., Schmaljohann, R., and Imhoff, J.F. 2011. Phylogenetic identification of fungi isolated from the Marine Sponge Tethya aurantium and identification of their secondary metabolites. Marine Drugs. 9: 561–585.
OPEN access freely available online
Chiang Mai University Journal of Natural Sciences [ISSN 16851994]
Chiang Mai University, Thailand
https://cmuj.cmu.ac.th
Muhammad Zainuddin1, Delianis Pringgenies1,*, Ocky Karna Radjasa1, 2 and Haeruddin3
1 Department of Marine Sciences, Faculty of Fisheries and Marine Sciences, Diponegoro University, Semarang, 50275, Indonesia
2 Deputy for Earth Sciences, Indonesian Institute of Sciences, Indonesia
3 Department Coastal Resources Management, Faculty of Fisheries and Marine Sciences, Diponegoro University, Semarang, 50275, Indonesia
Corresponding author: Delianis Pringgenies, E-mail: delianispringgenies@lecturer.undip.ac.id
Total Article Views
Editor: Korakot Nganvongpanit,
Chiang Mai University, Thailand
Article history:
Received: August 13, 2021;
Revised: September 27, 2021;
Accepted: October 4, 2021

