
Biodegradation of Microplastics by Microorganisms Isolated from Two Mature Landfill Leachates
Megga Ratnasari Pikoli*, Puji Astuti, Festy Auliyaur Rahmah, Arina Findo Sari, and Nur Amaliah SolihatPublished Date : 2022-01-21
DOI : https://doi.org/10.12982/CMUJNS.2022.005
Journal Issues : Number 1, January-March 2022
Abstract Microplastics are contaminants in the form of tiny plastic fragments diluted in terrestrial and aquatic environments. Recently, these contaminants have become a concern due to their negative impact on the quality of life of living things. The isolation and examination of microplastic degrading microorganisms' ability from two large mature landfills were conducted. Therefore, this study aims to obtain bacteria and fungi as bioremediation agents that can degrade microplastics. The isolation process was conducted by direct and indirect (enriched) methods. Nutrient agar and potato dextrose agar media were used either in the form of a full or a tenth of a recipe with the addition of polyethylene, polypropylene, and polystyrene microplastics. Furthermore, indirect isolation used mineral media treated with the same microplastics. Colony morphology was observed to be the difference among isolates. The isolates were selected based on their ability to produce lipase in butter agar, and their ability to use microplastic as the only carbon source was examined. A total of 211 isolates were obtained, consisting of 74 bacteria and 137 fungi. One-third of the total isolates produced lipase. A bacterial isolate with the highest lipase index identified based on the 16S rRNA gene showed that it was Bacillus paramycoides. The isolate used all three types of microplastics, with the highest ability in polystyrene, which was degraded up to 11.12% in 42 days. In conclusion, microorganisms isolated from the landfill leachate have potential as bioremediation agents that degrade microplastics.
Keywords: Bacillus, Bacillus paramycoides, Biodegradation, Bioremediation, Landfill, Leachate, Microplastics, Polystyrene
Funding: The authors are grateful for the research funding provided by the Center for Research and Publication UIN Syarif Hidayatullah Jakarta, Indonesia.
Citation: Pikoli, M.R., Astuti, P., Rahmah, F.A., Sari, A.F., and Solihat, N.A. 2022. Biodegradation of microplastics by microorganisms isolated from two mature landfill leachates. CMU J. Nat. Sci. 21(1): e2022005.
INTRODUCTION
Microplastics are environmental contaminants that have received necessary attention due to their increased quantity and persistence. It is restricted to small plastic pieces ranging from about 5 mm to a few micrometers in diameter (Andrady, 2017; Costa et al., 2018; Accinelli et al., 2019; Yuan et al., 2020; Tadsuwan and Babel, 2021). They are spread within the environment through atmospheric dust, rain, laundry waste, landfill sites, and wastewater treatment plant (Dris et al., 2018; Tadsuwan and Babel, 2021). A study showed that 275 million metric tons of plastic waste were generated by 192 countries in 2010. About 4.8 to 12.7 million metric tons were entering in the ocean, and it is estimated to double by 2025 due to lack of waste management (Jambeck et al., 2015).
Microplastics contamination in the environment results in disruption of the quality of life of living things, including humans. Predators at higher trophic levels in the food pyramid, such as humans are affected when it is ingested by tiny organisms (Andrady, 2017; Wu et al., 2017). Further consequences are pathological stress, reproductive complications, decreased growth rates, liver inflammation, and even microplastics absorbing and accumulating heavy metals from the surrounding environment (Auta et al., 2017). Most plastic debris spreads from land, but studies regarding microplastics in soil are still limited (Costa et al., 2018; He et al., 2018; Zhang et al., 2020).
Strategies to control microplastic contamination include: reducing it from the source, using biodegradable and compostable plastics, and developing remediation technologies to clean the environment. One of the possible and low-cost efforts of cleaning is bioremediation that utilizes the degradation capability of microorganisms. Although microplastics have high persistence, adapted microorganisms can still degrade them. Free-living microorganisms adapt to almost all environmental conditions, including transforming various polymers. Therefore, microplastic-degrading microorganisms can be obtained from an environment that has been polluted for a long time.
A study should be conducted on the degradation of microplastics by degrading microorganisms. Microplastic-degrading microorganisms increase natural biodegradation without damaging the environment. Therefore, this strategy is very promising for the bioremediation of natural ecosystems without significant side effects. Despite increasing publications and involving multidisciplinary collaborations, microplastic degradation biotechnology still requires a lot of research and various strategies due to limitations on highly stable plastic properties, environmental conditions, the ability of microorganisms themselves, and the influence of microplastic-microorganism interactions (Shen et al., 2019; Jiang et al., 2021; Zhang et al., 2021). Biotechnology starts from obtaining microorganisms, and research that isolates microplastic-degrading microbes is still limited; therefore, tough isolates of microorganisms still need to be explored to achieve great success and sustainability.
In current study, microorganisms were isolated from leachate that was filtered from waste in landfills. It was estimated that they had adapted to live with plastic materials as a source of nutrition and the environment. The isolation methods used were direct and indirect (enriched). Cipayung and Cipeucang landfills in Java Island were used as examples of where leachate was obtained. This is because they are one of the large mature landfills operating for decades. The isolates were selected for their ability to produce lipase. The selected isolate was assessed for its growth ability on microplastics and was identified for its species. Therefore, this study aims to obtain microplastic-degrading microorganisms and determine their degrading ability while using it as a sole carbon source.
MATERIALS AND METHODS
Sample, media, and chemicals
The sources of isolation were leachate from the Cipayung and Cipeucang landfills. The media used were nutrient agar (NA), nutrient broth (NB), potato dextrose agar (PDA), Bushnell-Haas (BH), Bushnell-Haas agar (BHA), microplastics of polyethylene (PE), polypropylene (PP), and polystyrene (PS), and butter agar. Molecular identification of isolate used DNA extraction kit, polymerase chain reaction (PCR) kit, primer 27F/1492R, agarose and buffer Tris-acetate EDTA pH 9.0. The PE, PP, and PS microplastics were prepared by cutting them into approximately 5x5 mm2. Furthermore, sterilization of the microplastics was carried out using ultraviolet lamp exposure. The BH medium contains magnesium sulphate 0,2 g/L, monopotassium phosphate 1 g/L, calcium chloride 0,02 g/L, ammonium nitrate 1 g/L, dipotassium phosphate 1 g/L, and ferric chloride 0,05 g/L. Butter agar contains peptone 5 g/L, yeast extract 2,5 g/L, agar 15 g/L, liquid butter 25 mL/L, tween 80 5 mL/L, and methylene blue 0,01 g/L.
Description of sampling sites
Leachates were sampled from 2 landfills, Cipayung and Cipeucang, each at three random sites. The sampling in the Cipayung landfill used codes CY1, CY2, and CY3, while CC1, CC2, and CC3 were used in the Cipeucang landfill. The three points were determined based on the difference in temperature of leachate measured on-site in the field (Table 1). Cipayung is located in Depok, West Java Province, while Cipeucang in South Tangerang, Banten Province, Indonesia. Then the physicochemical properties of the leachate samples were analyzed. Each landfill receives domestic waste from a district with a population of more than 2 million people. Both landfills are operated daily without intermediate covers and liners but have dumping arrangements.
Table 1. Description of the sampling sites in the Cipayung landfill and the Cipeucang landfill.
|
Landfill |
Altitude |
Coordinate |
Site |
Description |
Temperature (ºC) |
|
Cipayung |
93 feet |
6º19'29"S, 106º39'37"E |
CY1 |
Old waste pile |
25.2 ± 0.10 |
|
CY2 |
New waste pile |
31.23 ± 0.06 |
|||
|
CY3 |
Inlet of wastewater treatment |
33.5 ± 0.00 |
|||
|
Cipeucang |
325 feet |
6º25'18"S, 106º47'20"E |
CC1 |
Pipe A mix of old and new waste leachate |
30.7 ± 0.01 |
|
CC2 |
Pipe B mix of old and new waste leachate |
35.43 ± 0.06 |
|||
|
CC3 |
Inlet of wastewater treatment |
28.4 ± 0.01 |
Isolation of microorganisms
Direct and enriched isolations were the two main approaches used. In direct isolation, leachate samples were serially diluted by multiples of 10, then grown on NA and PDA media with a complete formula, and incubation was conducted at 28°C for two days. In addition, direct isolation was conducted by inoculation of leachate samples in general medium with 1/10 of the recipe, namely 1/10NA and 1/10PDA. Each plate was added with PE, PP, and PS microplastics without prior dilution of the sample, and incubation was carried out at the same temperature. Furthermore, the colonies growing around the microplastics were inoculated on fresh NA or PDA.
In enriched isolation, the leachate samples were enriched in BH medium, supplemented by 0.1% (w/v) of PE, PP, and PS microplastics; thoroughly shaken at 120 rpm at 28°C. After two weeks of incubation, when the turbidity of the medium became apparent, 1 mL of the sample was diluted and plated on some agar plates. The different colonies were observed, purified, and stored as slant agar cultures.
Selection of lipase-producing isolates
The isolates were selected by growing on butter agar which was stained with methylene blue. Every single type was inoculated onto 3 wells of 0.1 mL of the culture suspensions on the surface of butter agar plate, and the cultures were incubated in an incubator at 28°C for 48 hours. The diameter of the clear zone and colonies was measured using a micrometer, and the index was calculated against the colony diameter.
Examination of selected isolate
A seed culture was prepared in 30 mL of NB medium in a 100 mL Erlenmeyer flask by inoculating three loops of colonies from slanted agar. The incubation was carried out for 24 hours on a shaker at 100 rpm. The culture clarity on day 0 was documented, and at 24 hours, culture turbidity was checked. Successful growth occurred when it is cloudy. However, incubation was continued until the 48th hour when it is as clear as documented on day 0.
Inoculum culture was prepared in the following manner: 15 mL of the seed culture was poured onto 135 mL medium, which was a mixture of half formula NB and BH medium. The incubation was carried out for 24 hours on a shaker at a speed of 120 rpm.
A 45 mL of BH medium containing 0.1% (w/v) PE, PP, or PS microplastics were prepared with a total of 36 flasks for a sampling time of 7, 14, and 28, with three replicates and three kinds of microplastics. In addition, controls without microorganisms were made with the same amount. The liquid medium was sterilized by autoclaving before the addition of sterile microplastics.
Each test medium was then inoculated with 5 mL of inoculum culture aseptically, and incubation was carried out on a shaker at a speed of 120 rpm. At each sampling time, culture cultivation was stopped according to the planned time for harvesting microplastics. Furthermore, microplastics in the culture solution were filtered through an 80 Mesh wire filter. The entire solution, including microplastics, was poured onto the sieve, and the container was rinsed with water and filtered again to ensure that nothing remains in it. The collected microplastics were washed three times with distilled water, dried in an oven at 50°C, and weighed using an analytical balance. The degradation was calculated from the weight reduction of microplastics from the beginning to the end of the incubation period.
Identification of the selected isolate
The selected isolate was identified by a molecular method that PCR-amplified the 16S rRNA gene. The DNA was extracted with a spin-column bacterial extraction kit while being purified. Universal primer 27F/1492R was used for PCR with initial denaturation of 95°C for 10 minutes, denaturation of 95°C for 1 minute, annealing of 51°C for 1 minute, an extension of 72°C for 1 minute. The entire process was repeated for 30 cycles, then a final extension of 72°C for 10 minutes was conducted. The PCR product was confirmed by the appearance of a band measuring 1500 bp using agarose gel electrophoresis, and the product was sent for sequencing. Furthermore, the identity of the isolate was obtained from the bioinformatics analysis of the DNA sequence.
Data analysis
The leachate properties and the result on microorganism isolates were analyzed descriptively. Additionally, the results on selection and microplastics degradation were analyzed by analysis of variance to obtain significant difference among the treatments and continued by Duncan test (P < 0.05). The analysis of the selected isolate DNA sequence was carried out using bioinformatics. The quality of the DNA electropherogram was assessed qualitatively with the Embrapa program. The forward and reverse sequences were assembled, edited, and trimmed with ChromasPro (Technelysium Pty Ltd) to produce contig sequences. Sequence searches in GenBank that were homologous to contig were performed with BLASTn at https://blast.ncbi.nlm.nih.gov (Altschul et al., 1990). The search was limited to a 16S rRNA (bacteria and archaea) database of type material and optimized for very similar sequences (megablasts). After the 100 most similar sequences were produced, alignment was conducted with the MEGAX program (Kumar et al., 2018). Furthermore, the reconstruction of the phylogenetic tree used the Neighbor-Joining method with bootstrap 1000 times. The outgroup used was the 16S rRNA sequence of Escherichia coli, and the treeGraph2 program enhanced the resulting phylogenetic tree view. The sequence was registered to GenBank after the previous checking whether they include chimeras through DECIPHER version 2.17.1 (Wright et al., 2012).
RESULTS
Physicochemical properties of leachate samples
The three samples from the sampling points at the two landfills had differences in temperature and pH (Table 1). This showed the difference in content and properties as confirmed by the phytochemical results (Table 2). The leachate from the old waste pile in the Cipayung landfill (CY1) has a moderate pH and temperature, as well as the lowest sulfate and ammonia since it was neutralized by the environment. Meanwhile, the new waste pile leachate (CY2) has a lower pH, high total dissolved solids, iron, biological oxygen demand, chemical oxygen demand, and total suspended solids. The leachate from the inlet wastewater treatment Cipayung (CY3) was a mixture of the old and new waste piles, so the physicochemical properties are a combination of the two wastes.
Meanwhile, CC1 and CC2 are leachates from a mixture of old and new waste with different pipes. They have relatively different temperatures, pH, sulfate, zinc, iron, turbidity, total suspended solids, and ammonia. The leachate from the inlet wastewater treatment of Cipeucang (CC3) was also very different in properties, incredibly high in sulfate, total dissolved solids, KMnO4, cadmium, chemical oxygen demand, and ammonia.
Table 2. Physicochemical characteristics of the Cipayung landfill and Cipeucang landfill leachate samples.
|
Properties |
Sample code |
|||||||||||
|
CY1 |
CY2 |
CY3 |
CC1 |
CC2 |
CC3 |
|
||||||
|
pH |
9.13 |
7.38 |
9.04 |
9.39 |
8.75 |
9.15 |
||||||
|
Temp (°C) |
25 |
26 |
25 |
31 |
35 |
35 |
||||||
|
BOD (mg/L) |
493.887 ± 21.351 |
800.746 ± 29.002 |
585.049 ± 30.594 |
250.647 ± 22.420 |
262.423 ± 21.763 |
491.033 ± 21.659 |
||||||
|
COD (mg/L) |
2873.344 ± 99.923 |
4653.568 ± 161.831 |
3123.200 ± 108.612 |
1188.096 ± 0.000 |
1188.096 ± 0.000 |
2424.934 ± 8.617 |
||||||
|
Turbidity (NTU) |
254 |
389 |
233 |
743 |
56 |
100 |
||||||
|
TSS (mg/L) |
188 |
306 |
219 |
174 |
62 |
75 |
||||||
|
TDS (mg/L) |
5950 |
10025 |
7775 |
6300 |
6275 |
13100 |
||||||
|
Ammonia (NH3-N mg/L) |
572.670 ± 0.656 |
871.874 ± 0.295 |
969.834 ± 3.456 |
9982.997 ± 5.657 |
24566 ± 45.962 |
129277.49 ± 156.978 |
||||||
|
Sulfate (mg/L) |
1.05 |
11.43 |
7.43 |
15.99 |
1.31 |
2549 |
||||||
|
Oil and fat (mg/L) |
0.55 |
0.3 |
0.35 |
0.1 |
0.06 |
0.06 |
||||||
|
KMnO4 (mg/L) |
50.9 |
52.8 |
30.7 |
39.5 |
47.1 |
75.5 |
||||||
|
Iron (mg/L) |
2.991 ± 0.005 |
14.713 ± 0.008 |
4.541 ± 0.068 |
6.249 ± 0.034 |
1.887 ± 0.021 |
3.432 ± 0.006 |
||||||
|
Copper (mg/L) |
0.0076 ± 0.001 |
0.041 ± 0.002 |
0.026 ± 0.003 |
0.04 ± 0.04 |
0.034 ± 0.013 |
0.010 ± 0.004 |
||||||
|
Lead (mg/L) |
0.119 ± 0.000 |
0.049 ± 0.000 |
<0.040 |
0.500 ± 0.152 |
0.370 ± 0.040 |
0.138 ± 0.009 |
||||||
|
Zinc (mg/L) |
0.250 ± 0.002 |
0.448 ± 0.001 |
0.188 ± 0.000 |
0.546 ± 0.011 |
0.022 ± 0.003 |
0.121 ± 0.003 |
||||||
|
Cadmium (mg/L) |
<0.006 |
<0.006 |
<0.006 |
<0.002 |
<0.002 |
0.002 |
||||||
Note: Temp= temperature, BOD= Biological oxygen demand, COD= Chemical oxygen demand, TSS= Total suspended solids, TDS= Total dissolved solids
Obtained isolates
Isolation from landfill leachate was conducted by direct and indirect methods to capture as many bacteria as possible that can be cultured. The recapitulation of the number obtained from the two landfills can be seen in Table 3. Bacteria isolated from the Cipayung landfill from the direct method using the complete formula of NA medium were six types per sampling point.
Table 3. The number of isolates obtained from Cipayung dan Cipeucang landfills.
|
Organisms |
Media used |
Cipayung Landfill |
Cipeucang Landfill |
Number of isolates |
||||||
|
CY1 |
CY2 |
CY3 |
CC1 |
CC2 |
CC3 |
|||||
|
Bacteria |
NA |
6 |
6 |
6 |
10 |
6 |
6 |
40 |
74 |
|
|
1/10NA+microplastics |
1 |
10 |
8 |
3 |
3 |
0 |
25 |
|||
|
BH+microplastics |
0 |
2 |
2 |
3 |
2 |
0 |
9 |
|||
|
Fungi |
PDA |
4 |
3 |
9 |
6 |
14 |
17 |
53 |
137 |
|
|
1/10PDA+microplastics |
14 |
10 |
15 |
4 |
3 |
3 |
49 |
|||
|
BH+microplastics |
8 |
3 |
5 |
11 |
5 |
3 |
35 |
|
||
|
|
Total isolates |
211 |
||||||||
Meanwhile, bacteria isolated from the Cipeucang landfill through the direct method with complete formula NA medium amounted to 6-10 kinds per sampling site. The number obtained from a mixture of old and new waste leachate (CC1) was the highest from the Cipeucang landfill. This was considered related to the mild physicochemical characteristics compared to other sampling sites, which is favorable for most heterotrophic bacteria that can grow on an NA medium. Furthermore, the types of bacterial isolates (40 isolates) obtained from the two landfills, Cipayung and Cipeucang, were isolated mainly by the direct method on the complete formula of NA medium. The efforts to obtain specific bacteria were carried out using 1/10NA and BH medium, each with the addition of PE, PP, and PS microplastics. Through this approach, additional 25 and 9 bacterial isolates, respectively, were obtained.
Isolation of fungi from landfill leachate was conducted by varying the medium and methods, like in bacteria isolation, to capture as many fungi as possible. The types of fungi from leachate inlet wastewater treatment (CY3 or CC3) were more diverse than those from the other two sampling sites. This fact was consistent with its nature, a mixture of leachates from old and new waste piles. Therefore, the combination of physicochemical properties from the two sites in the same landfill makes the content diverse as micro-nutrients. Direct isolation of fungi using 1/10PDA general medium from the Cipayung landfill obtained more varieties, especially from leachate of old waste piles (CY1). However, this does not apply to the results of direct isolation of fungi using 1/10PDA medium from the Cipeucang landfill, which yields less than those using the full formula of PDA.
Results on lipase-producer selection
Not all isolates obtained were viable at the time of inoculum preparation for the selection of lipase producers. A total of 164 viable isolates from 211 in the Cipayung and Cipeucang landfills were selected for their ability to produce lipase. This selection was based on the presence of a clear zone on the butter agar. The index formed described the ability to produce lipase semi-quantitatively. Also, the halo zone on the butter agar medium showed the butter or lipids degraded by lipases secreted by microorganisms.
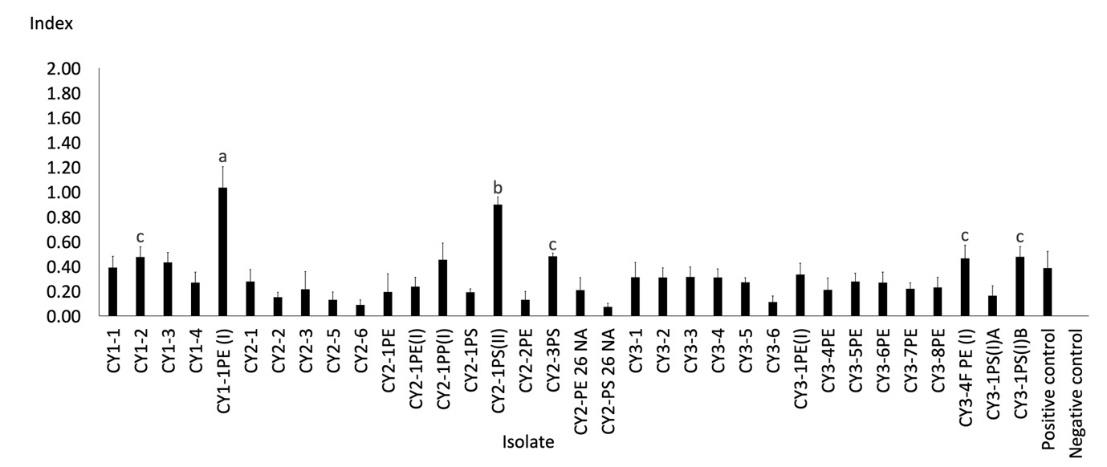
Figure 1. Clear zone index from the lipase-producing test of Cipayung landfill isolates.
Note: The different letters above the bars indicate a significant difference from the Duncan test results (P < 0.05)
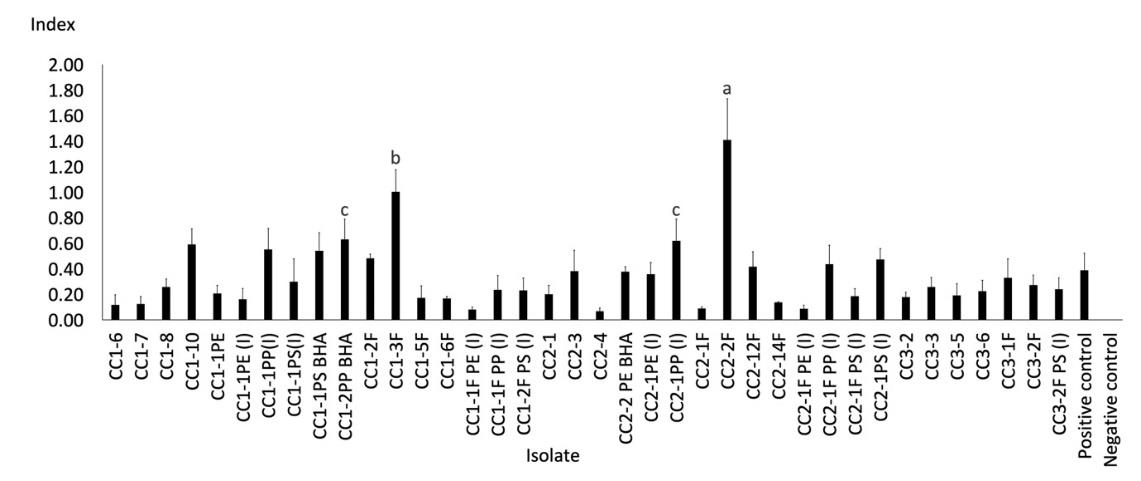
Figure 2. Clear zone index from the lipase-producing test of Cipeucang landfill isolates.
Note: The different letters above the bars indicate a significant difference from the Duncan test results (P < 0.05).
The selection results showed that lipase was produced by 72 isolates, where 34 were from Cipayung (Figure 1) and 38 from Cipeucang (Figure 2). This result means that at least one-third of the total isolates obtained from the landfill are lipase producers because of their potential to become microplastic-degrading agents. The isolates that showed the most extensive clear zone index of lipase activity were bacteria CY1-PE(I) from the Cipayung landfill and the fungi CC2-2F from the Cipeucang landfill. The two isolates, and other isolates that are also having the ability to produce lipase, have the potential to be developed as microplastic-degrading agents.
Microplastics degradation by the selected isolate
The isolate assessed for its degradation activity in the present study was CY1-1PE(I), selected from several other isolates, which also showed great potential. This Cipayung landfill’s isolate was chosen because of its ease of growth. It was isolated by the direct method using 1/10NA medium with the addition of PE microplastic after incubation for seven days. The percentage of degradation (Figure 3) was consistent with bacteria isolation when the culture was seven days old. On the 7th day of the degradation activity test, PE microplastics' degradation percentage was higher than those of PP and PS. The difference was more visible on the 14th day than before. However, on day-28 and 42, the percentage of PS microplastics degradation was greater than PE, which jumped to 7.14% and 11.12%, respectively. Therefore, CY1-1PE(I) has the highest potential to degrade PS microplastics.
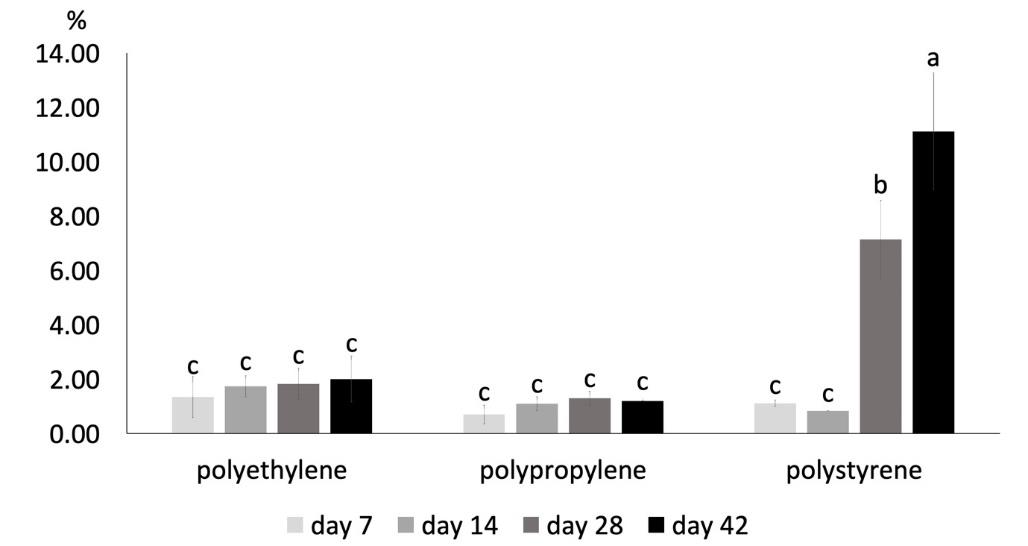
Figure 3. Percentage of microplastics degradation by CY1-1PE(I).
Note: The different letters above the bars indicate a significant difference from the Duncan test results (P < 0.05)
Identity of isolate CY1-1PE(I)
The CY1-1PE(I) was identified based on the 16S rRNA gene fragment analysis. The phylogenetic tree (Figure 4) confirmed the bacterial identity as Bacillus paramycoides, which had 100% similarity with Bacillus subtilis strain MCCC 1A04098 (Liu et al., 2017). The 16S rRNA gene sequence of Bacillus paramycoides CY1-1PE(I) was deposited in the NCBI GenBank and had an accession number MT995121 (https://www.ncbi.nlm.nih.gov/nuccore/MT995121).
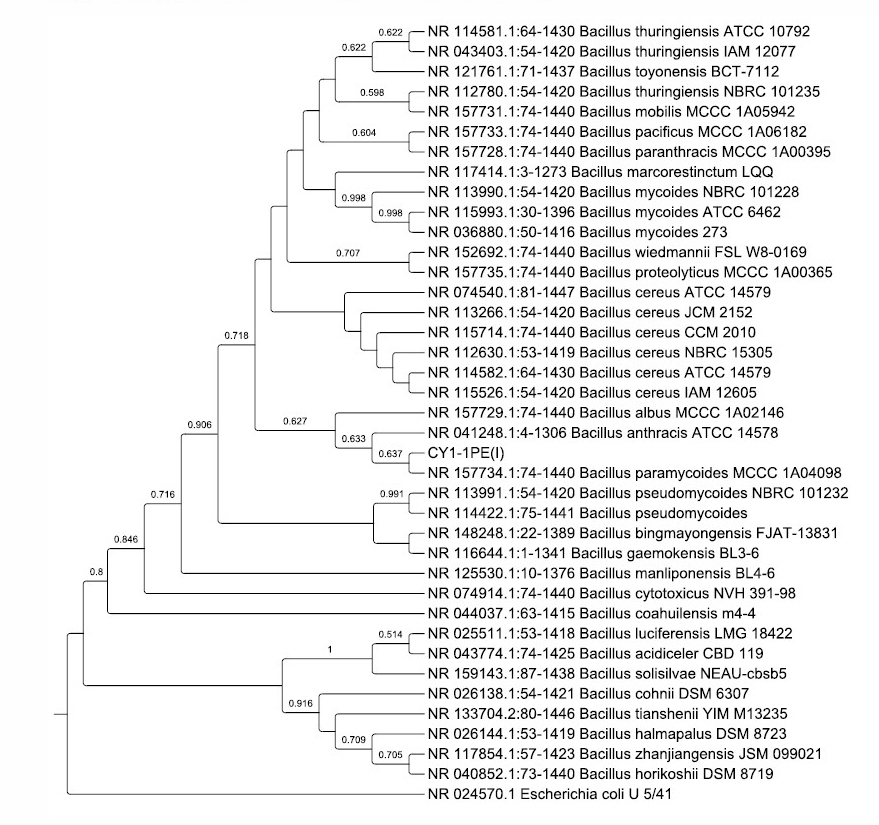
Figure 4. Phylogenetic tree of CY1-1PE(I) isolates and their closest homologous sequences based on 1367 base pairs of the 16S rRNA gene
DISCUSSION
Each sampling site in both landfills has its own physicochemical properties that allow different microorganisms to be obtained. It is known that conventionally treated wastewater releases up to 2.32 X 109 microplastic particles per day (Tadsuwan and Babel, 2021). The presence of microplastics in such an effluent creates an adapting factor in the microbial community that inhabits it. Leachate of Cipayung landfill was sampled from the old and new waste piles, while in the Cipeucang landfill, the old waste is mixed with the new waste so that the sampling was only distinguished from the leachate flow sites, which were pipe A and B. However, in both landfills, sampling was carried out on the liquid from the inlet or leachate entering the wastewater treatment. The differences in physicochemical properties of all sampling points showed the potential for obtaining different microorganisms, both in diversity and activity, even in the same landfill. The distribution of microorganisms was neither uniform nor random but depended on physical, chemical, and biological processes and varies on spatial and temporal scales (Lew et al., 2019).
The bacteria obtained from the new waste pile leachate (CY2) had the largest average cell concentration compared to the other two sampling sites (data not shown). It was consistent with the highest biological oxygen demand value in the CY2 sample, which described the high content of easily degradable organic matter in the sample, and compatibility with the moderate temperature and pH. High biological oxygen demand described the number of heterotrophic aerobic microorganisms in the sample. Although this study did not assess the feasibility of leachate characteristics, the value of the biological oxygen demand of all samples was high (more than 150 mg/L) based on environmental regulations regarding the quality standards of raw landfill leachate (Regulation of the Minister of Environment and Forestry Republic of Indonesia No. P.59/Menlhk/Setjen/Kum.1/7/2016, 2016).
Meanwhile, bacteria obtained from the Cipeucang landfill was less than those from the Cipayung. Nevertheless, with high sulfate and ammonia conditions, it is expected that the bacteria from this sample were adapted to these conditions. High concentrations of ammonia and sulfate allowed the formation of ammonium sulfate. In other experiments that provide high ammonium sulfate, limited glucose increased the surface hydrophobicity of Pseudomonas sp. AKS2 on polyethylene plastic. The bacteria were initially reared for three days at 30°C. Furthermore, they were given polyethylene plastic and incubated for another 12 days at the same temperature (Tribedi & Sil, 2014).
It is not easy to determine whether they were repeated or identical isolates because the colony morphology showed its distinct characteristics in each isolate. This number is thought to represent 0.1% of the similar group of bacteria present in the leachate sample. The cultivation and isolation of microorganisms by conventional methods only obtain 0.1-1.0% of soil bacteria (Daniel, 2005). Therefore, it is predicted that there are various kinds of bacteria in the Cipayung and Cipeucang landfill leachates from the heterotrophic bacteria group, which cannot be isolated (unculturable). It is a challenge to obtain the bacteria that are difficult to culture.
The purpose of using the 1/10 recipe medium was to capture more slow-growing microorganisms. In general, the complete formula of the NA medium is rich in nutrients, which benefits fast-growing microorganisms and ignores the slow-growing. Therefore, many studies have succeeded in growing previously unculturable bacteria from various aquatic and terrestrial habitats using the diluted medium (Vartoukian et al., 2010).
The isolation also used an indirect method with liquid BH medium, which was then grown on BH agar plates. The medium was a mineral basalt with a carbon source of microplastics. The use of microplastics is expected to be a selector for microplastic-using bacteria. Enrichment media select bacteria by growing the targeted bacteria while eliminating the nontargeted (Bonnet et al., 2020). Even though the enrichment was considered right on target, it could not isolate all of them. Therefore, the gain was less compared to the direct method.
Isolation of fungi was carried out using three kinds of media as well. The average cell concentration of individual fungi from CY3 and CC3 was lower than to those from the other sites (data not shown), whereas it is well known that the abundance of microorganisms can increase when the content of organic matter and nutrients in the environment increases (Siles and Margesin, 2016). It is because although diverse communities have greater access to resources, average individual taxa within such communities have lower resource availability and carbon use efficiency; competition and complementation increase simultaneously with diversity up to a threshold level (Yu et al., 2019). Although their growth was limited, it is expected that the fungi isolated from this sites will adapt to microplastics in landfill waste. According to a review by Yuan et al. (2020), various kinds of fungi are widely distributed in the environment that show high reproductive ability in using microplastics as a carbon source.
The presence of organic content in the 1/10PDA medium allowed the isolation of organic carbon users. Similarly, the presence of microplastics in the medium allowed the isolation of microplastics users. The content of both PDA and microplastics, allowed the isolation of heterotrophic fungi adapted to the environment with microplastics and use more gradually over time. The isolation of fungi was complemented by an indirect method strategy, which also used BH and microplastics as a medium. The BH medium with microplastics captured users of plastic materials as the only carbon source. The use of microplastics, especially those enriched in the indirect method, caused the growth of microorganisms to be selected. It is similar to another study, in situ enrichment experiments in the estuary and marine environments with microplastics for 14 days to form specific communities (Oberbeckmann et al., 2018). Therefore, direct isolation with full and 1/10 formula medium complemented the obtained fungal isolates, plus those obtained from the enrichment method with BH and microplastics medium. Other studies also adopted this strategy, such as isolating polyurethane-degrading fungi from landfills using a minimal medium, PDA, and a combination of the media (Magnin et al., 2018).
Lipase is used as a selection factor because the biodegradation of polymers by bacteria and fungi is considered to be played by lipases (Shah et al., 2016a; Yoshida et al., 2016). This group of enzymes has broad substrate specificity that can degrade various polymers such as plastics, plant oils, triglycerides, and fatty acid methyl esters (Ghosh et al., 2013). Furthermore, most of the waste in landfills comes from households, including waste oils and fats from plants and animals; and plastic packaging. These materials become adapting factors of microorganisms since they can be used as substrates.
Another interesting finding is that they have the potential either as a single culture or in a consortium culture. For example, isolate CY3-1PS(I)B as the third-largest lipase index from the Cipayung landfill has a symbiont CY3-1PS(I)A, which showed a lower clear zone index (Figure 1). Their abilities are expected to increase as their growth are combined. The ability of degradation can be increased when the microorganism is present as a consortium culture (Shah et al., 2016b).
The percentage of degradation of 11.12% after 42 days is a great ability compared to bacteria from other studies. Bacillus cereus degraded PP microplastic by 1.6% and PS by 7.4% within 40 days (Auta et al., 2017). Exiguobacterium sp. derived from the digestive tract of Tenebrio molitor worm larvae degraded pieces of polystyrene by 7.4% for more than 60 days (Ho et al., 2018). Although it showed promising potential, the use of CY1-1PE(I) in degrading microplastics should be improved to become faster and have a much higher percentage of degradation.
The identification of CY1-1PE(I) as Bacillus paramycoides indicates that this isolate will have the ability as a bioremediation agent. Many species of the genus Bacillus are known as microplastic degrading bacteria from various studies. These species include Bacillus amyloliquefaciens (Das & Kumar, 2015), Bacillus cereus dan Bacillus gottheilii (Auta et al., 2017), Bacillus subtilis (Shah et al., 2016a), Bacillus brevis (Ghosh et al., 2013), and other Bacillus sp. (Park & Kim, 2019; Yuan et al., 2020). Their tough nature in dealing with unusual environmental conditions with various xenobiotics compounds causes them to be often used in bioremediation.
Bacillus paramycoides strain MCCC 1A04098 as the closest to CY1-1PE(I) was proposed as a novel species from the Bacillus cereus group. This strain was isolated from the South China Sea sediments. Strains within the Bacillus cereus group shared more than 97% and less than 95% similarity with other Bacillus species. This claim is based on a polyphasic taxonomic approach of biochemical tests and analysis of 16S rRNA gene sequences. The members of the group B. cereus are facultative anaerobes, form spores, and are widely distributed in various environments (Liu et al., 2017).
The strains of Bacillus paramycoides were published regarding their potential in the bioremediation of xenobiotics. A strain of Bacillus paramycoides from other study showing 99.86% homology with strain MCCC 1A04098 was considered a potential agent for bioremediation of wastewater from domestic, textile, hospital, drug manufacturing, and mixed wastes (Rashid et al., 2020). Three strains of Bacillus paramycoides were isolated from petroleum-contaminated soil in Iraq (Hamzah et al., 2020). Meanwhile, another strain of Bacillus paramycoides was combined in consortium culture with Bacillus paranthracis for bioremediation of soil contaminated with fluoranthene, a pollutant polycyclic aromatic hydrocarbons (PAHs) in agricultural land (Wang et al., 2020).
The biodegradation ability of bacteria has been proven on various substrates, even on recalcitrant substrates, such as xenobiotics. Bacteria, especially those that mobile and free-living, are cosmopolitan and develop adaptive abilities to live in various environments with the addition of materials that are no longer natural. This ability can be exploited by making them biotechnological agents, especially bioremediation. By obtaining the potential isolates in this study, the mature leachate landfill was confirmed to be an abundant source of microbes for harvesting microplastic-degrading agents.
CONCLUSION
Microorganisms (bacteria and fungi) were isolated from the leachate in the Cipayung and the Cipeucang landfills, approximately one-third of which produced lipase and therefore have the potential to degrade microplastics. The assessed isolate, CY1-1PE(I) bacteria, showed the highest ability to degrade polystyrene microplastics, with a degradation percentage of 11.12% for 42 days. Furthermore, it was identified as Bacillus paramycoides based on 16S rRNA gene sequence analysis. Future studies should be conducted to further assess fungal and other potential isolates as a single culture or as a consortium.
ACKNOWLEDGEMENTS
The authors appreciate the Center for Research and Publication UIN Syarif Hidayatullah Jakarta for the financial support under grant number UN.01/KPA/455/2020 and UN.01/KPA/455/2021, and to UPTD Laboratorium Lingkungan Dinas Lingkungan Hidup Kota Tangerang Selatan for measuring of the physicochemical properties of the leachates.
AUTHOR CONTRIBUTIONS
Megga Ratnasari Pikoli brought the main idea, conducted sampling and experiments, performed the data analysis and visualization, and wrote the manuscript. Puji Astuti, Festy Auliyaur Rahmah, Arina Findo Sari, and Nur Amaliah Solihat handled samples, conducted the experiments, contributed to statistical analysis and the manuscript writing. All authors have read and approved the final manuscript.
CONFLICT OF INTEREST
The authors declare that they hold no competing interests.
REFERENCES
Accinelli, C., Abbas, H.K., Shier, W.T., Vicari, A., Little, N.S., Aloise, M.R., and Giacomini, S. 2019. Degradation of microplastic seed film-coating fragments in soil. Chemosphere. 226: 645–650.
Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. Journal of Molecular Biology. 215: 403–410.
Andrady, A.L. 2017. The plastic in microplastics : A review. Marine Pollution Bulletin Journal. 11: 12–22.
Auta, H.S., Emenike, C.U., and Fauziah, S.H. 2017. Screening of Bacillus strains isolated from mangrove ecosystems in Peninsular Malaysia for microplastic degradation. Environmental Pollution Journa. 231: 1552–1559.
Bonnet, M., Lagier, J.C., Raoult, D., and Khelai, S. 2020. Bacterial culture through selective and non-selective conditions : The evolution of culture media in clinical microbiology. New Microbes and New Infections. 34: 1–11.
Costa, J.P., Paco, A., Santos, P.S.M., Duarte, A.C., and Rocha-santos, T. 2018. Microplastics in soils: Assessment, analytics and risks. Environmental Chemistry. 16: 18–30.
Daniel, R. 2005. The metagenomics of soil. Nature Reviews. Microbiology. 3: 470–478.
Das, M.P. and Kumar, S. 2015. An approach to low-density polyethylene biodegradation by Bacillus amyloliquefaciens. 3 Biotech. 5: 81–86.
Dris, R., Gasperi, J., and Tassin, B. 2018. Sources and fate of microplastics in urban areas: A focus on Paris megacity. In M. Wagner & S. Lambert (Eds.), Freshwater microplastics: Emerging environmental contaminants? (pp. 69–83). Springer Open.
Ghosh, S.K., Pal, S., and Ray, S. 2013. Study of microbes having potentiality for biodegradation of plastics. Environmental Science Pollution Research. 20: 4339–4355.
Hamzah, A.F., Al-Tamimi, W.H., Mahdi, S.S., and Alameri, N.Z. 2020. Isolation and identification new bacterial strains isolated from different sources of Al-Rafidiyah oil field in Iraq. CATRINA, 21: 15–22.
He, D., Luo, Y., Lu, S., Liu, M., Song, Y., and Lei, L. 2018. Microplastics in soils: Analytical methods, pollution characteristics and ecological risks. Trends in Analytical Chemistry. 109: 163–172.
Ho, B.T., Roberts, T.K., and Lucas, S. 2018. An overview on biodegradation of polystyrene and modified polystyrene: The microbial approach. Critical Reviews in Biotechnology. 38: 308–320.
Jambeck, J.R., Geyer, R., Wilcox, C., Siegler, T.R., Perryman, M., Andrady, A., Narayan, R., and Law, K.L. 2015. Plastic waste inputs from land into the ocean. Science. 347: 768–771.
Jiang, Y., Xia, W., Zhao, R., Wang, M., Tang, J., and Wei, Y. 2021. Insight into the interaction between microplastics and microorganisms based on a bibliometric and visualized analysis. Bulletin of Environmental Contamination and Toxicology. 1–12.
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. 2018. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution. 35: 1547–1549.
Lew, S., Glińska-Lewczuk, K., and Lew, M. 2019. The effects of environmental parameters on the microbial activity in peat-bog lakes. PLoS ONE. 14: e0, 1–18.
Liu, Y., Du, J., Lai, Q., Zeng, R., Ye, D., Xu, J., and Shao, Z. 2017. Proposal of nine novel species of the Bacillus cereus group. International Journal of Systematic and Evolutionary Microbiology. 67: 2499–2508.
Magnin, A., Hoornaert, L., Pollet, E., and Phalip, V. 2018. Isolation and characterization of different promising fungi for biological waste management of polyurethanes. Microbial Biotechnology. 12: 544–555.
Regulation of the Minister of Environment and Forestry Republic of Indonesia No. P.59/Menlhk/Setjen/Kum.1/7/2016, 1 (2016).
Oberbeckmann, S., Kreikemeyer, B., Labrenz, M., and Harrison, J.P. 2018. Environmental factors support the formation of specific bacterial assemblages on microplastics. Frontiers in Microbiology. 8: 1–12.
Park, S.Y., and Kim, C.G. 2019. Chemosphere Biodegradation of micro-polyethylene particles by bacterial colonization of a mixed microbial consortium isolated from a land fill site. Chemosphere. 222: 527–533.
Rashid, A., Mirza, S.A., Keating, C., Ali, S., Campos, L.C. 2020. Indigenous Bacillus paramycoides and Alcaligenes faecalis : Potential solution for the bioremediation of wastewaters. BioRxiv.
Shah, Z., Gulzar, M., Hasan, F., and Shah, A.A. 2016a. Degradation of polyester polyurethane by an indigenously developed consortium of Pseudomonas and Bacillus species isolated from soil. Polymer Degradation and Stability. 134: 349–356.
Shah, Z., Gulzar, M., Hasan, F., and Shah, A.A. 2016b. Degradation of polyester polyurethane by an indigenously developed consortium of Pseudomonas and Bacillus species isolated from soil. Polymer Degradation and Stability. 134: 349–356.
Shen, M., Zeng, G., Zhang, Y., Wen, X., Song, B., and Tang, W. 2019. Can biotechnology strategies effectively manage environmental (micro)plastics? Science of the Total Environment. 697: 134200.
Siles, J.A., and Margesin, R. 2016. Abundance and diversity of bacterial, archaeal, and fungal communities along an altitudinal gradient in Alpine forest soils: What are the driving factors? Microbial Ecology. 72: 207–220.
Tadsuwan, K., and Babel, S. 2021. Microplastic contamination in a conventional wastewater treatment plant in Thailand. Waste Management and Research. 39: 754–761.
Tribedi, P., and Sil, A.K. 2014. Cell surface hydrophobicity : A key component in the degradation of polyethylene succinate by Pseudomonas sp. Journal of Applied Microbiology. 116: 295–303.
Vartoukian, S.R., Palmer, R.M., and Wade, W.G. 2010. Strategies for culture of “unculturable” bacteria. FEMS Microbiology Letters. 309: 1–7.
Wang, M., Zhang, H., and Li, G. 2020. Biodegradation of fluoranthene contaminated soil by different combinations of bacteria biodegradation of fluoranthene contaminated soil by different combinations of bacteria. IOP Conference Series: Earth and Environmental Science. 526: 1–4.
Wright, E.S., Yilmaz, L.S., and Noguera, D.R. 2012. DECIPHER, a search-based approach to chimera identification for 16S rRNA sequences. Applied and Environmental Microbiology. 78: 717–725.
Wu, W., Yang, J., and Criddle, C.S. 2017. Microplastics pollution and reduction strategies. Frontiers of Environmental Science & Engineering. 11: 1–4.
Yoshida, S., Hiraga, K., Takehana, T., Taniguchi, I., Yamaji, H., Maeda, Y., Toyohara, K., Miyamoto, K., Kimura, Y., and Oda, K. 2016. A bacterium that degrades and assimilates poly (ethylene terephthalate). Science. 351: 1196–1199.
Yu, X., Polz, M.F., and Alm, E.J. 2019. Interactions in self-assembled microbial communities saturate with diversity. The ISME Journal. 13: 1602–1617.
Yuan, J., Ma, J., Sun, Y., Zhou, T., Zhao, Y., and Yu, F. 2020. Microbial degradation and other environmental aspects of microplastics/plastics. Science of the Total Environment. 715: 136968.
Zhang, B., Yang, X., Chen, L., Chao, J., Teng, J., and Wang, Q. 2020. Microplastics in soils: A review of possible sources, analytical methods and ecological impacts. Journal of Chemical Technology and Biotechnology. 95: 2052–2068.
Zhang, X., Li, Y., Ouyang, D., Lei, J., Tan, Q., Xie, L., Li, Z., Liu, T., Xiao, Y., Farooq, T. H., et al. 2021. Systematical review of interactions between microplastics and microorganisms in the soil environment. Journal of Hazardous Materials. 418: 126288.
OPEN access freely available online
Chiang Mai University Journal of Natural Sciences [ISSN 16851994]
Chiang Mai University, Thailand
https://cmuj.cmu.ac.th
Megga Ratnasari Pikoli1,*, Puji Astuti2, Festy Auliyaur Rahmah2, Arina Findo Sari1, and Nur Amaliah Solihat2
1 Department of Biology Faculty of Science and Technology Universitas Islam Negeri (UIN) Syarif Hidayatullah Jakarta, Indonesia
2 Center for Integrated Laboratory Universitas Islam Negeri (UIN) Syarif Hidayatullah Jakarta, Indonesia
Corresponding author: Megga Ratnasari Pikoli, E-mail: meggapikoli@uinjkt.ac.id
Total Article Views
Editor: Supon Ananta,
Chiang Mai University, Thailand
Article history:
Received: July 19, 2021;
Revised: September 3, 2021;
Accepted: September 13, 2021

