
Different Responses of ESBL Indicative Peptide Spectra to Various β- Lactam Exposures Among Community Acquired Urinary Tract Infected Escherichia coli by Using the MALDI-TOF Technique
Siraphob Lananta, Pontawat Siriratanagool, Nakarin Sommanawan, Pattadon Lerttrakarnnon, Sararun Boonchuay, Supavit Jirawattanapong, Sirinya Manochomphu, Thanapat Sastraruji, and Siriwoot Sookkhee*Published Date : 2021-09-13
DOI : https://doi.org/10.12982/CMUJNS.2021.095
Journal Issues : Number 4, October-December 2021
Abstract The study aims to identify Escherichia coli specific and Extended Spectrum β- Lactamase (ESBL) indicative peptide spectra when co-exposed with ceftazidime or other beta-lactams with clavulanic acid in ESBL producing isolates in community acquired urinary tract infected E. coli by Matrix-Assisted Laser Desorption/Ionization Time - of - Flight Mass Spectrometry (MALDI-TOF) technique. Among 100 tested E. coli isolates, 13 antibiotics were used to profile the resistant isolates. They were mainly demonstrated the resistance to ampicillin, cefoxitin, cefotaxime, ceftazidime, amoxicillin/clavulanic acid, and tetracycline. The high-, low-, and non-ESBL producers, 43.86%, 40.35% and 15.79%, were classified from these resistant isolates according to the ratio of MIC fold between ceftazidime and amoxicillin/clavulanic acid by E-test. Using the MALDI-TOF technique, 5 E. coli -specific peptide spectra were identified which were located at 4362, 4531, 5380, 6254, and 9063 Da using a cut-off value of 60% of frequency. The significant intensity reduction of spectra at 10477, and 5096 Da were recorded and suspected as ESBL indicative peptide spectra after the exposure to ceftazidime or cefotaxime alone and concomitantly with amoxicillin/clavulanic acid in the high and low ESBL producers, respectively. The further significant spectra located at 2548, and 8371 and 9713 Da were exclusively related to ceftazidime hydrolysis in the low and high ESBL producers, respectively. There were 3 and 4 significant peptide spectra which were located at 5968, 7153, 9713 and 10477 Da, and 2548, 5096, and 9537 Da were also suspected as being ESBL spectra after exposure to other tested β-lactams in the high and low-ESBL producers, respectively.
Keywords: Extended Spectrum β-Lactamase; E. coli; MALDI-TOF, Peptide spectra; β-lactams
Funding: The authors are thankful for the research funding provided by the Faculty of Medicine, Chiang Mai University, Chiang Mai, Thailand.
Citation: Lananta, S., Siriratanagool, P., Sommanawan, N., Lerttrakarnnon, P., Boonchuay, S., Jirawattanapong, S., Manochomphu,S., Sastraruji, T., and Sookkhee, S. 2021. Different responses of ESBL indicative peptide spectra to various β- Lactam exposures among community acquired urinary tract infected Escherichia coli by using the MALDI-TOF technique. CMU J. Nat. Sci. 20(4): e2021095.
INTRODUCTION
Bacterial resistance is one of the most common causes of death worldwide. In 2014, over 700,000 people died from bacterial infection and the number could reach 10 million annually by the end of year 2050 (Kraker et al., 2016). The incidence of antibiotic-resistant bacterial infection in Thailand is also over 88,000 cases per year (Sumpradit et al., 2017). One of the most prevalent infections among Thais is the community acquired urinary tract infection caused by Escherichia coli which is the most prevalent antibiotic-resistant bacteria (Flores-Mireles et al., 2015). Normally, the process used to make a conventional diagnosis of the causative agent in bacterial infection still takes a long time, 3-5 days, resulting in extensive progression of the infection. This may result in ineffective treatment particularly if empirical drugs or broad-spectrum drugs have been inactivated by bacterial enzymes and their use if less effective. Inappropriate use of antibiotics causes an increase in drug resistance in bacteria. This is a major factor causing antibiotic resistance globally (Melander et al., 2018). Hence, antibiotic resistance needs to be addressed if we are to shorten the length of identification and susceptibility tests as well as improving drug prescription. The capability to generate a β-lactamase enzyme is the crucial mechanism in causing resistance in E. coli. The β-lactamase enzyme is responsible for the hydrolysis of an amide bond in the β-lactam ring inactivating the efficacy of cephalosporins and penicillins. Extended-spectrum β-lactamase (ESBL), a member of class-A β-lactamases, is the most common class to inactivate the members of β-lactams namely the penicillins, monobactams, and third generation cephalosporins (Picozzi et al., 2014).
VITEK-MS, an automated mass spectrometry microbial identification system, uses Matrix-Assisted Laser Desorption / Ionization Time - of - Flight Mass Spectrometry (MALDI-TOF MS) techniques to perform the bacterial susceptibility by displaying bacterial peptide spectra according to their masses. This apparatus allows scientists to identify a causative agent within a minute (Singhal et al., 2015). It also benefits laboratory scientists as it can be used to detect a variety of drug-resistant proteins as peptide spectra. In medical treatment, VITEK-MS can benefit infectious disease physicians as it improves accurate diagnosis enabling swift, effective treatment improving the survival rate. In the present study, we aimed to investigate the peptide spectra of community acquired uropathogenic Escherichia coli which were exposed to various β-lactams using MALTI-TOF techniques to enable the identification of ESBLs that could possibly be the cause of antimicrobial resistance.
MATERIALS AND METHODS
Bacterial samples
One hundred isolates of community acquired uropathogenic E. coli were collected between January and April, 2019 from the Diagnostic Laboratory Unit, Maharaj Nakorn Chiang Mai Hospital, Faculty of Medicine, Chiang Mai University and recovered on MacConkey agar plates. An isolated colony grown on each plate was collected for further characterization and biochemical identification. The recovered isolates were inoculated separately in Tryptic Soy Broth supplemented with 20% glycerol, then kept at -20°C until investigation at the Department of Microbiology, Faculty of Medicine, Chiang Mai University. This present study was approved for biosafety by the Faculty of Medicine, Chiang Mai University.
Identification of the collected bacteria
E. coli gives pink colonies on MacConkey agar and produces A/AG on Triple Sugar Iron agar as well as indole production. The Triple Sugar Iron slant (TSI) technique was also performed a change in color from red to yellow showing the production of carbon dioxide gas in anaerobic condition at the bottom which were caused by E. coli growth. E. coli can generate carbon dioxide in anaerobic condition and convert tryptophan to indole. E. coli is also able to generate acid condition and catalase resulting in positive results in the case of methyl red and catalase tests. E. coli is also further confirmed by the ability to convert glucose to acetone (positive Voges-Proskauer test), urea to ammonia and carbon dioxide (positive urease test). These isolates were also identified as E. coli after confirmation by Vitek-MS. E. coli ATCC25922 was used as the standard control bacteria.
Antibiotic susceptibility test
In this study, each isolate was tested with 13 antibiotics (Oxoid, Basingstoke, UK) including ampicillin (AMP, 30 µg/ml), amoxicillin-clavulanic acid (AMC, 30/10 µg/ml), imipenem (IPM, 30 µg/ml), cefoxitin (FOX, 30 µg/ml), ceftazidime (CAZ, 30 µg/ml), cefotaxime (CTX, 30 µg/ml), piperacillin/tazobactam (TZP, 30/10 µg/ml), ciprofloxacin (CIP, 30 µg/ml), tetracycline (TE, 30 µg/ml), gentamicin (CN, 30 µg/ml), kanamycin (K, 30 µg/ml), and meropenem (MEM, 30 µg/ml), and doripenem (DOR, 30 µg/ml). E. coli isolates that were susceptible to less than or equal to 7 drugs were counted as resistant strains while the intermediate and susceptible strains were susceptible to a range of 8-9 and 10-13 drugs, respectively. A single colony of each isolate was inoculated into Mueller Hinton Broth (MHB; Oxoid, Basingstoke, UK) before the turbidity being adjusted to equal McFarland standard No. 0.5, then evenly swabbed on Mueller Hinton Agar (MHA; Oxoid, Basingstoke, UK) plates. The disks containing the antibiotics described above were placed on the plates using a disk dispenser. Then the cultures were incubated at 37°C for 18-24 hours. The diameter of the inhibition zone in millimeters was measured and interpreted in accordance with CLSI guidelines (CLSI, 2018).
Detection of extended spectrum β- lactamase
ESBL production was screened using the double disc diffusion technique on the Mueller Hinton agar (Naseer et al., 2017). An AMC disc was placed on swabbed plates 20 mm from the CTX disc and then incubated at 37°C for 18-24 hours. The inhibition zone and keyhole phenomenon of each disc were recorded and interpreted for susceptibility to CTX. This assay was also done with CAZ disc. If the inhibition zone was ≤ 22 mm (in the case of CTX) or ≤ 27 mm (in the case of CAZ) or keyhole phenomena were observed, it was marked as an ESBL production.
High and low ESBL production were categorized according to the MIC ratios of the ESBL E-strip test (Sookkhee et al, 2017). Firstly, each freshly prepared suspension was separately swabbed onto MHA plates. Then, an E- strip (bioMérieux™, Marcy l'Etoile, France). which contains a gradient concentration of AMC with CAZ at one end and CAZ at the opposite end were placed on the agar. After 37°C incubation for 24 hours, MIC values were determined. Ratio between MICCAZ and MICCAZ/AMC were calculated. Low and high ESBL productions were determined in accordance with MIC ratios between 8 and 250, and ≥ 250-fold, respectively. Conversely, no ESBL production was indicated if the MIC ratio was below 8.
Slide preparation for peptide determination via VITEK-MS
Each β- lactam tested was placed on an MHA plate after swabbing each tested isolate and incubated at 37°C. Two exposed area including the area which was exposed to a tested β- lactam (arrow 1), and the area which exposed to a tested β- lactam and amoxicillin-clavulanic acid (arrow 2) as shown in Figure. 1 were performed for the peptide determination.
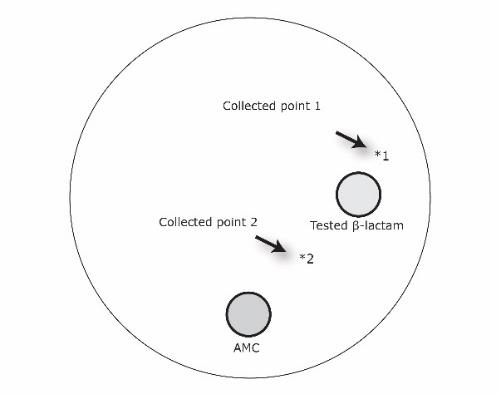
Figure 1. Collection points of the culture isolate after exposure to the tested β- lactam alone (point 1), and the tested β-lactam and amoxicillin-clavulanic acid (point 2).
A single colony of each isolate was picked up from the 2 exposed areas and smeared separately on a well of VITEK MS-DS target slides (bioMérieux™, Marcy l'Etoile, France). The center of each acquisition spot group has a calibrator spot used to detect normal function of the VITEK -MS. E. coli ATCC8739 was applied to calibrator spot as control strain. Alpha-4-cyano-4-hydroxycinnamic acid (CHCA) matrix (bioMérieux™, Marcy l'Etoile, France) was then dropped on top of each well to give crystallization. VITEK-MS (bioMérieux™, Marcy l'Etoile, France) was run on a laser frequency of 50 Hz with wavelength 337 nm, ion source voltage 20 kV and lens voltage 6 kV to produce peptide spectra. The peptide spectra were presented as mass in Dalton unit (Da) and spectrum intensity.
Peptide spectra analysis
Peptide spectra from each well of the target slide were determined. In this study, a frequency peptide spectrum higher than 70% from the total collected sample point as described above was calculated and marked as suspected E. coli specific spectra. To analyze the peptide spectra responsible for ESBL production, MALDI/TOF-MS technique was used to investigate the relationship between peptide spectra and the susceptibility to various β-lactams in the high, low, and non ESBL-producing E. coli. Importantly, ESBL- producing E. coli should demonstrate a statistically significant difference of peptide spectra after exposure to clavulanic acid. The same peptide spectra would help to identify the significant differences in spectra in ESBL production exposed to various β- lactams and clavulanic acid as described above in the double disc diffusion.
Statistical analysis
Between the exposure of the tested β- lactam alone (point 1) compared with the exposure of the tested β- lactam and amoxicillin-clavulanic acid (point 2), the reduction in peptide intensity and its level of significance at the identical peptide mass was determined. The peptide spectra which showed a significant reduction may be suspected as being ESBL indicative spectra. The similarity and differences in of ESBL indicative spectra which resulted from the exposure of each tested β- lactams were also determined. The significant differences in peptide spectra were then calculated among various β- lactams, the presence of keyhole phenomenon and ESBL production. Paired sample T-test was performed to analyze the significant difference between the spectrum intensity at point 1 and point 2. The software package used was the Statistical Package for Social Sciences (SPSS version 17.0 Inc., Chicago, IL), and the differences were considered significant when p-values were less than 0.05.
RESULTS
Categorization of E. coli according to the antibiotic susceptibility
Of the 100 isolates of E. coli tested after determination of the susceptibility to 13 antibiotics, 57%, 16% and 27% were categorized as resistant, intermediate resistant and susceptible isolates which were susceptible to ≤7, 8-9, and 10-13 antibiotics, respectively.
From the experiments, the percentage resistance of total isolates was much higher than those susceptible when tested against AMP, FOX, CTX, CAZ, AMC and TE (85 and 15%, 72 and 28%, 85 and 15%, 79 and 21%, 76 and 24%, 76 and 24% respectively). However, the percentage resistance of total isolates was slightly greater than percentage susceptibility when exposed to CIP (66 and 34%). Conversely, when the samples were tested against IPM, TZP, MEM, DOR, K, CN, the percentage of susceptible samples were significantly higher than the percentage of those which were resistant. On exposure to IPM, the percentage of susceptible E. coli was approximately 94%. Whilst when tested with K, three-quarters of the samples were susceptible. The results were more noticeable when the samples were tested against TZP, MEM, DOR and CN as the samples were totally susceptible to these drugs (100%), suggesting that although most of the antibiotics are resisted by E. coli, there are still these effective drugs for the treatment of E. coli infection.
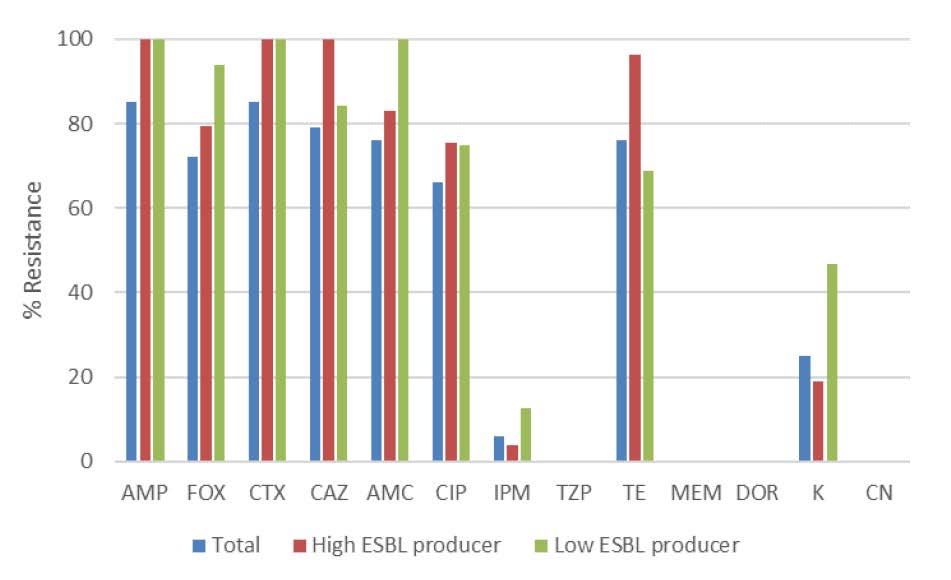
Figure 2. Antimicrobial susceptibility of the E. coli strain tested. AMP, ampicillin; AMC, amoxicillin-clavulanic acid; IPM, imipenem; FOX, cefoxitin; CAZ, ceftazidime; CTX, cefotaxime; TZP, piperacillin/tazobactam; CIP, ciprofloxacin; TE, Tetracycline; MEM, meropenem; DOR, doripenem; K, kanamycin; CN, gentamicin.
Detection of extended spectrum β-lactamase
In the double disk synergy test, the ESBL production of E. coli could be detected from the presence of keyhole phenomenon and the zone of clearance around the CAZ and CTX disks. Fifty six out of 57 isolates (98.25%) in the resistant group were suspected as ESBL-producing E. coli. According to the E-strip test results of the resistant isolates, 43.86% of isolates were classified as high ESBL- producing E. coli which exhibited a ratio of MIC value ≥ 250-fold. Low ESBL- producing E. coli accounted for 40.35% which exhibited a ratio of MIC value between 8- to 250-fold. While in 15.79% of them, the ratio of MIC ≤ 8-fold were then interpreted as being non-ESBL-producers.
Peptide spectra analysis
Among the identification of E. coli, 6 E. coli-specific peptide spectra were identified by using VITEK-MS including spectra located at 4362, 4531, 5096, 5380, 6254, and 9063 Da. Three peptide spectra, 4531, 5380, and 6254 Da were detected in the range of 50-60% between the non and high ESBL- producing groups. Data are shown in Table 1.
Apart from E. coli specific spectra after exposure to CAZ alone, and CAZ with AMC, 6 spectra were identified as being significant as shown in Table 2. Three of these, located at 8371, 9713, and 10477 Da, were significantly and exclusively noted as being in a high ESBL-producing group. It may be assumed that these peptide spectra were responsible for ESBL-indicative peptide spectra in a high ESBL-producing group whereas only one spectrum, 2548 Da was an ESBL- indicative peptide spectrum in a low ESBL-producing group. Two peptide spectra located at 2875 and 6314 Da were assumed to be susceptible peptide spectra due to their sole appearance in the non ESBL-producing group of E. coli.
Table 1. E. coli specific peptide spectra analyzed from VITEK-MS that exhibits in the high, low, and non ESBL-producing groups.
|
Peptide spectra located at (Da) |
Percentage frequency of peptide spectra detected |
|||
|
Total isolates |
High ESBL group |
Low ESBL group |
Non ESBL group |
|
|
4362 |
85.33 |
69.23 |
91.91 |
91.41 |
|
4531 |
73.10 |
74.04 |
85.29 |
59.38* |
|
5380 |
74.18 |
59.62* |
72.79 |
87.50 |
|
6254 |
70.92 |
50.00* |
79.41 |
78.91 |
|
9063 |
82.61 |
73.07 |
93.38 |
78.91 |
Table 2. Peptide spectra which showed a significant difference after exposure to CAZ alone, CAZ concomitant with AMC.
|
ESBL groups |
Spectra (Da) |
Mean ± SE |
Significant difference (P) |
||
|
CAZ exposed |
Clavulanic exposed |
|
|||
|
High
|
8371 |
245,498 ± 175,697 |
338,088 ± 171,586 |
0.037* |
|
|
9713 |
136,655 ± 59,252 |
89,970 ± 34,512 |
0.008* |
||
|
10477 |
53,576 ± 33,111 |
72,279 ± 28,536 |
0.029* |
||
|
Low
|
2548 |
12,191 ± 2,482 |
22,110 ± 2,515 |
0.002* |
|
|
5096 |
81,591 ± 31,338 |
55,211 ± 27,340 |
0.038* |
||
|
Non
|
2875 |
56,179 ± 29,794 |
36,789 ± 24,867 |
0.022* |
|
|
6314 |
36,334 ± 16,214 |
45,126 ± 16,989 |
0.042* |
||
Note: *P-value < 0.05.
After exposure to CTX alone, and CTX with AMC, a peptide spectrum located at 10477 Da was significantly shown in the high ESBL- producing E. coli but only one spectrum at 5380 Da was detected among the non ESBL- producing group as shown in Table 3. There was only one spectrum which was found to be significant in the low ESBL- producing group after exposure to CTX alone, and CTX concomitant with AMC. It is located at 5096 Da but is not mentioned as being in the ESBL- indicative spectra due to it was categorized as being in the E. coli-specific spectrum as described above.
Table 3. Peptide spectra which demonstrated significant differences after exposure to CTX alone, and CTX concomitant with AMC.
|
ESBL groups |
Spectra (Da) |
Mean ± SE |
Significant difference (P) |
|
|
CAZ exposed |
Clavulanic exposed |
|||
|
High
|
5096# |
93,111 ± 44,964 |
56,063 ± 38,045 |
<0.001* |
|
10477 |
51,594 ± 36,758 |
62,657 ± 30,783 |
0.047* |
|
|
Low |
5096# |
81,591 ± 31,338 |
55,211 ± 27,340 |
0.001* |
|
Non |
5380 |
87,653 ± 31,012 |
82,538 ± 34,698 |
0.022* |
Note: #Spectrum categorized as E. coli specific peptide spectra as described above; *P-value < 0.05.
From the results shown above, it may be suggested that the high ESBL- producing E. coli shared a common peptide spectrum located at 10477 Da after exposure to either CAZ or CTX. It may be suggested that the peptide spectrum at 10477 Da plays a role as an ESBL enzyme for the hydrolysis of CAZ, and CTX. However, 2 peptide spectra which are located at 8371 and 9713 Da may be also responsible as specifically hydrolysis agents of CAZ in the high ESBL- producing group.
Further analysis of the significant differences in peptide spectra among the 7 other tested β-lactams, specifically AMP, FOX, CTX, CAZ, AMC, IPM, TZP, is required. These significant spectra are shown in Table 4. Due to the high ESBL- production of the tested E. coli and their resistance to AMP, CTX, CAZ, and AMC, the shared peptide spectra located at 5968, 7153, 9713, and 10477 Da at the significant P-values (P = 0.036, 0.030, 0.001, and 0.009, respectively) after exposure to these agents need further analysis. It may be suggested that these significant spectra were responsible as ESBL enzymes for hydrolyzing such antibiotics in conditions of high ESBL- production. Additionally, four remarkable peptide spectra located at 5968, 7153, 9713, and 10477 Da were also statistically significant in the high ESBL- producing group according to E-strip tests (P = 0.036, 0.030, 0.001, and 0.009, respectively) but only spectrum 9713 Da was only significant at P = 0.001 in the isolates which exhibited the keyhole phenomenon after the double disc diffusion test. Hence, it may be suggested that the 4 peptide spectra located at 5968, 7153, 9713, and 10477 Da are significant in the high ESBL- producing group, which can interrupt the action of β-lactams. On the contrary, these spectra were also significantly detected according to susceptibility to IMP and TZP in the high ESBL-producing E. coli. Only one spectrum, 9537 Da, was exclusively shown to be significant in a high ESBL-producing group after exposure to FOX (P = 0.044), which may suggest this is a specific ESBL which causes the resistance to FOX.
According to the results, low ESBL E. coli shared common peptide spectra located at 2538 and 9537 Da at significant P-values (P = 0.002, and 0.032, respectively) after exposure to AMP, FOX, CAZ, CTX, AMC as shown in Table 5. The peptide spectrum located at 2548 Da was significantly detected when tested against TZP and IMP (P = 0.002, and 0.002, respectively). A significant spectrum located at 9537 Da was also evident in the keyhole detected group.
Table 4. Significant differences in peptide spectra found in the high ESBL- producing group after analysis of the antimicrobial susceptibility of 7 β-lactams, keyhole phenomenon and ESBL production.
|
|
Significant difference (P) of peptide spectra (Da) |
||||||||
|
2690 |
4362 |
4531 |
5380 |
5968 |
7153 |
9537 |
9713 |
10477 |
|
|
AMP |
0.508 |
0.129 |
0.399 |
0.056 |
0.036* |
0.030* |
0.270 |
0.001** |
0.009* |
|
FOX |
0.508 |
0.043* |
0.484 |
0.027* |
ND |
0.089 |
0.044* |
0.001** |
0.009* |
|
CTX |
0.508 |
0.129 |
0.399 |
0.056 |
0.036* |
0.030* |
0.270 |
0.001** |
0.009* |
|
CAZ |
0.508 |
0.129 |
0.399 |
0.056 |
0.036* |
0.030* |
0.270 |
0.001** |
0.009* |
|
AMC |
0.508 |
0.045* |
0.604 |
0.056 |
0.036* |
0.030* |
0.270 |
0.001** |
0.007* |
|
IMP |
0.508 |
0.210 |
0.010** |
0.175 |
0.036* |
0.030* |
0.270 |
<0.001** |
0.012* |
|
TZP |
0.020* |
0.472 |
0.088 |
0.034* |
ND |
0.087 |
0.044* |
0.007* |
0.027* |
Note: ND, not detected; *, P < 0.05; **, P ≤ 0.001
Table 5. Significant differences of peptide spectra demonstrated in the low ESBL- production group after analysis of the antimicrobial susceptibility of 7 β-lactams.
|
|
Significant difference (P) of peptide spectra located at (Da) |
||
|
2548 |
9537 |
9713 |
|
|
AMP |
0.002* |
0.032* |
0.138 |
|
FOX |
0.002* |
0.032* |
0.138 |
|
CTX |
0.002* |
0.032* |
0.138 |
|
CAZ |
0.002* |
0.032* |
0.007* |
|
AMC |
0.002* |
0.032* |
0.138 |
|
IMP |
0.002* |
0.080 |
0.17 |
|
TZP |
0.002* |
0.080 |
0.197 |
Note: ND, not detected; *, P < 0.05
From the results above, we focused on a common peptide spectrum located at 10477 Da after exposure to either CAZ or CTX. It may play a role as ESBL in the hydrolysis of CAZ, and CTX and other β lactams except TZP.
To analyze the significance of this spectrum compared with CTX susceptibility, it was found to be significant in the resistant groups of AMP, FOX, CAZ, CTX, and AMC as shown in Table 6. It may be suggested that a spectrum located at 10477 Da plays an essential role as an ESBL among low ESBL groups. Conversely, this spectrum was more significant in the susceptible group rather than the resistant group when tested with IMP and TZP.
Table 6. Significant differences in peptide spectra demonstrated in the high ESBL production group after analysis of the antimicrobial susceptibility of 7 β-lactams, keyhole phenomenon and ESBL production compared with CTX.
|
|
Significant difference (P) of peptide spectra located at (Da) |
|||
|
4365 |
4532 |
7274 |
10477 |
|
|
AMP |
0.165 |
0.313 |
0.134 |
0.022* |
|
FOX |
0.048* |
0.313 |
0.134 |
0.022* |
|
CTX |
0.165 |
0.313 |
0.134 |
0.022* |
|
CAZ |
0.165 |
0.313 |
0.134 |
0.022* |
|
AMC |
0.064 |
0.474 |
0.134 |
0.017* |
|
IMP |
0.497 |
0.266 |
ND |
ND |
|
TZP |
0.964 |
0.506 |
0.632 |
ND |
Note: *, P < 0.05
DISCUSSION
As conventional susceptibility tests could not provide answers regarding any antibiotic resistant proteins, we explored advanced molecular and proteomic techniques to enable identification of these antibiotic resistant genes or proteins. These complicated techniques are also expensive and require highly experienced operators. Hence, to date these modern in vitro diagnostic (IVD) methods have not been applied in routine laboratory diagnoses in developing countries such as Thailand. Currently, VITEK-MS is routinely carried out to identify the causative agents in the rapid and more precise daily laboratory diagnosis by using the MALDI-TOF technique. VITEK-MS is easier to practice, less time-consuming and is cheaper per sample unit. VITEK-MS can analyze peptide spectra within a minute and it also has high specificity and sensitivity (Miller et al., 2018; Arumugham and Cascella, 2020; M Américo et al., 2020). Many reports have shown that VITEK-MS is routinely used to identify the causative agents by displaying species-specific peptide spectra (Rychert et al., 2013).
Of the peptide spectra produced from the VITEK-MS process, a lot of remaining peptide spectra are always ignored despite the fact that they might indicate some significant proteins in the bacterial cells. The reason we chose to study the peptide spectra in E. coli is this is the most common causative pathogen in antibiotic resistance as well as being a common uropathogenic infection. Although it is the most common causative agent, there are few reports relating to the peptide spectra of the proteins causing antibiotic resistance in E. coli resulting in limited range of knowledge applicable to laboratory diagnosis. Currently, there is only a previous study that reported details peptide spectra of carbapenemases (Lasserre et al., 2015). Whereas, there is no evidence related to the analysis the peptide spectra of ESBLs. However, our laboratory research group are focusing on the identification of peptide spectra related to ESBLs which have been collected from the different population of various ESBL-producing bacteria.
We are interested in studying regarding ESBLs due to them being associated with most common mechanisms in antibiotic resistance of E. coli. They can hydrolyze penicillins, cephalosporins and monobactams but not carbapenems. ESBLs, which hydrolyze β-lactam rings in cephalosporins and penicillins, play a major role in antibiotic resistance among the majority of E. coli strains (Saka et al., 2020). However, we were unable to indicate whether E. coli produced the same ESBL against each antibiotic agent. These actions could be expressed by the identical or different characteristics of peptide spectra in the case of each antibiotic, which were produced from the same and distinct species of ESBL.
In this study, cefotaxime and ceftazidime were used as different representatives of third generation cephalosporins to investigate whether different ESBLs were produced when treated with different antibiotics. Different peptide spectra from non-identical antibiotics can be useful in the diagnosis of specific ESBLs which cause the resistance to each antibiotic. Being able to identify peptide spectra of ESBLs would help to reduce incidence of resistance in the third generation cephalosporins and penicillins.
Even though there were studies related to ESBLs in the past, we believe that there are changes in population, mechanisms, and the variety of ESBLs as time progresses. Moreover, we had planned to use third generation cephalosporins drugs (ceftazidime and cefotaxime) rather than one drug to observe a wider range of ESBL activity. Hence, we believe this study would generate strong evidence to represent peptide spectra of current ESBLs.
Identification of bacteria was carried out by a variety of biochemical tests. This increased the reliability of the results when confirming the samples as E. coli. A wide range of drugs were used in susceptibility tests. This increased confidence in the results of resistant, intermediate resistant and susceptible strains. Species-specific spectra were identified using VITEK-MS. Six species-specific peptide spectra were detected as being specific to E. coli, which corresponds to the findings published in a previous report (Wang et al., 2014).
Results of our study suggest that the peptide spectra located at 8371, 9713, and 10477 Da, and at 2548 Da were responsible as ESBL enzymes after exposure to ceftazidime alone and in combination with clavulanic acid among high and low ESBL producing E. coli. There was only one significant spectrum that could be detected in a high ESBL group after exposure to both ceftazidime and cefotaxime. This spectrum was located at 10477 Da. Moreover, peptide fingerprints located at 8371 and 9713 Da that were only expressed in high ESBL- producing E. coli after exposure to ceftazidime may be responsible as the specific ESBL for hydrolyzing ceftazidime.
In this study, peptide spectra were also determined after the tested isolates were exposed to different β-lactams in addition to ceftazidime and cefotaxime. This was done to help us to consider whether there are a variety of ESBLs in the response of E. coli to different antibiotics.
Results showed that 4 significant spectra which were located at 5968, 7153, 9713, and 10477 Da were commonly detected after the tested isolates were exposed to these 5 different antibiotics including ampicillin, cefoxitin, cefotaxime, ceftazidime and amoxicillin/clavulanate. It may be suggested that these spectra may play a role as ESBLs which produced among the high ESBL-producing E. coli. Additionally, only one peptide spectrum located at 9537 Da was detected and concluded as being a specific ESBL in response to cefotaxime exposure.
The results were substantiated when these 4 peptide spectra were not significantly detected when E. coli was exposed to imipenem and piperacillin/tazobactam. This is because imipenem is able to kill E. coli while piperacillin/tazobactam can interrupt the activity of the ESBL enzyme. Hence, ESBL activity decreased resulting in undetectable significant peptide spectra. These results support the assumption that the peptide spectra located 5968, 7153, 9713 and 10477 Da are the ESBLs in a high ESBL-producing E. coli. On the other hand, low ESBL- producing E. coli shared common peptide spectra located at 2538, and 9537 Da, finding which were statistically significant after being exposed to ampicillin, cefoxitin, ceftazidime, cefotaxime and amoxicillin/clavulanic acid.
The peptide spectrum located at 10477 Da was suspected of playing an essential role as an ESBL in the low ESBL- group as well despite being commonly found among ESBL E. coli. Nevertheless, the results in the low ESBL group showed there was no significant peptide spectra. Apart from the E. coli specific spectra, peptide spectra located at 9539, and 10463 Da were not significantly related to ESBL production in the low ESBL group. Nevertheless, the results among the low ESBL group showed that there were no significant peptide spectra (data not shown). There was no significant peptide spectrum detected when isolates were exposed to IMP and TZP in both the high and low ESBL groups. This is because TZP and IMP are not substrates of ESBLs. Hence ESBL is not generated when the samples were exposed to these antibiotics. Differences in detected peptide spectra between these β-lactams indicate that there are a variety of ESBLs playing roles in resistance to other β- lactams.
CONCLUSION
Although ceftazidime and cefotaxime are the members of third generation cephalosporins, their peptide spectra generated by E. coli were different after analysis using VITEK MS. It may be suggested that the different ESBL productions were dependent on the exposure of each drug. Therefore, the history of prescription for each patient was also affected by the identification of the peptides which were responsible for antibiotic resistance by using MALDI-TOF. Accurate identification of the ESBL peptide spectra could help to rapidly and more precisely identify β-lactam resistance. It would enable rapid prescription of the appropriate antibiotic prescriptions by the physician, which would potentially help to reduce the infection rate, mortality rate, and budgets and increase the viability of the antibiotics which remain effective against these bacteria which cause such detriment to our patients.
ACKNOWLEDGEMENTS
The authors sincerely thank to our technicians at Diagnostic Laboratory Unit, Maharaj Nakorn Chiang Mai Hospital, Faculty of Medicine, Chiang Mai University, Chiang Mai, Thailand for providing the bacterial samples and Vitek MS, and Department of Microbiology, Faculty of Medicine, Chiang Mai University, Chiang Mai, Thailand, for all the support and use of their facilities.
AUTHOR CONTRIBUTIONS
Siraphob Lananta wrote the manuscript. Pontawat Siriratanagool, Pattadon Lerttrakarnnon, and Sararun Boonchuay collected the Vitek MS spectra. Nakarin Sommanawan did the bacterial experiment. Supavit Jirawattanapong read the final manuscript. Sirinya Manochomphu collected the tested bacteria and operated Vitek MS. Thanapat Sastraruji performed statistical analysis. Siriwoot Sookkhee designed the proposal and experiments, did the bacterial experiment, wrote the manuscript. All authors have read and approved of the final manuscript.
LIMITATION AND SUGGESTION
For further investigation, there should be a study into the resistance of, not only E. coli, but also other ESBL producing bacterial species to the other drugs using MALDI-TOF technique. Eventually, in long-terms, a large resistant peptide spectra database of any bacteria should be generated and could be used as an indicator for faster diagnosis and more precise antibiotic prescription in the future.
CONFLICT OF INTEREST
The authors declare that they hold no competing interests.
REFERENCES
Arumugham, V.B., and Cascella. M. 2020. Third generation cephalosporins. Statpearls. Treasure Island (FL): StatPearls Publishing Copyright© 2020, StatPearls Publishing LLC.
Clinical and Laboratory Standards Institute (CLSI). 2018. Performance Standards for Antimicrobial Susceptibility Testing. 28th ed. CLSI supplement M100. Wayne, Pennsylvania, USA
Flores-Mireles, A.L., Walker, J.N., Caparon, M., and Hultgren, S.J. 2015. Urinary tract infections (Epidemiology, mechanisms of infection and treatment options). Nature Reviews Microbiology. 13: 269-284.
Kraker, M., Stewardson, A.J., and Harbarth, S. 2016. Will 10 million people die a year due to antimicrobial resistance by 2050?. PLOS Medicine. 13: e1002184.
Lasserre, C., De Saint Martin, L., Cuzon, G., Bogaerts, P., Lamar, E., Glupczynski, Y., Naas, T., and Tandé, D. 2015. Efficient detection of carbapenemase activity in Enterobacteriaceae by matrix-assisted laser desorption ionization− time of flight mass spectrometry in less than 30 minutes. Journal of Clinical Microbiology. 53: 2163-2171.
M Américo, F., P Machado Siqueira, L., B Del Negro, G.M., M Favero Gimenes, V., S Trindade, M. R., L Motta, A., Santos de Freitas, R., Rossi, F., L Colombo, A., Benard, G., et al. 2020. Evaluating VITEK MS for the identification of clinically relevant Aspergillus species. Medical Mycology. 58:322-327.
Melander, R.J., Zurawski, D.V., and Melander, C. 2018. Narrow-spectrum antibacterial agents. Medicinal Chemistry Communications. 9: 12-21.
Miller, E., Cantrell, C., Beard, M., Derylak, A., Babady, N.E., McMillen, T, Miranda E., Body B., Tang Y., Vasireddy R., et al. 2018. Performance of VITEK MS v3. 0 for identification of Mycobacterium species from patient samples by use of automated liquid medium systems. Journal of Clinical Microbiology. 56: e00219-18.
Naseer, F., Iqbal, R., Ikram, N., Shoaib, M., Javaid asad, M., Mehmood, RT., et al. 2017. Phenotypic cofirmatory disc diffusion test (PCDDT), double disc synergy test (DDST), E-test OS diagnostic tool for detection of extended spectrum beta lactamase (ESBL) producing Uropathogens. Journal of Applied Biotechnology and Bioengineering. 3:344-349.
Picozzi, S.C., Casellato, S., Rossini, M., Paola, G., Tejada, M., and Costa, E. 2014. Extended-spectrum β-lactamase-positive Escherichia coli causing complicated upper urinary tract infection (Urologist should act in time). Urology Annals. 6: 107-112.
Rychert, J., Burnham, C.A., Bythrow, M., Garner, O.B., Ginocchio, C.C., Jennemann, R., Lewinski, M.A., Manji, R., Mochon, A.B., Procop, G.W., et al. 2013. Multicenter evaluation of the VITEK MS matrix-assisted laser desorption ionization–time of flight mass spectrometry system for identification of Gram-positive aerobic bacteria. Journal of Clinical Microbiology. 51:225-2231.
Saka, H.K., García-Soto, S., Dabo, N.T., Lopez-Chavarrias, V., Muhammad, B., Ugarte-Ruiz, M., and Alvarez, J. 2020. Molecular detection of extended spectrum β-lactamase genes in Escherichia coli clinical isolates from diarrhoeic children in Kan. Nigeria. PloS ONE. 15: e0243130.
Singhal, N., Kumar, M., Kanaujia, P.K., and Virdi, J.S. 2015. MALDI-TOF mass spectrometry: an emerging technology for microbial identification and diagnosis. Frontiers in Microbiology. 6:791.
Sookkhee, S., Khantawa, B., and Srihinkong, W. 2017. Proteomic analysis of DNA starvation/stationary phase protection proteins from extended spectrum β-lactamase producing Escherichia coli. Chiang Mai University Journal of Natural Sciences. 16: 215-230.
Sumpradit, N., Wongkongkathep, S., Poonpolsup, S., Janejai, N., Paveenkittiporn, W., Boonyarit, P., Jaroenpoj, S., Kiatying-Angsulee, N., Kalpravidh, W., and Sommanustweechai, A. 2017. New chapter in tackling antimicrobial resistance in Thailand. British Medical Journal. 358: 20-24.
Wang, W., Xi, H., Huang, M., Wang, J., Fan, M., Chen, Y., Shao, H., and Li, X. 2014. Performance of mass spectrometric identification of bacteria and yeasts routinely isolated in a clinical microbiology laboratory using MALDI-TOF MS. Journal of Thoracic Disease. 6: 524–533.
OPEN access freely available online
Chiang Mai University Journal of Natural Sciences [ISSN 16851994]
Chiang Mai University, Thailand
https://cmuj.cmu.ac.th
Siraphob Lananta1, Pontawat Siriratanagool1, Nakarin Sommanawan1, Pattadon Lerttrakarnnon1, Sararun Boonchuay1, Supavit Jirawattanapong1, Sirinya Manochomphu2, Thanapat Sastraruji3, and Siriwoot Sookkhee1,*
1 Department of Microbiology, Faculty of Medicine, Chiang Mai University, Chiang Mai, Thailand 50200, Thailand.
2 Diagnostic Laboratory Unit, Maharaj Nakorn Chiang Mai Hospital, Faculty of Medicine, Chiang Mai University, Chiang Mai, Thailand 50200, Thailand.
3 Dental Research Center, Faculty of Dentistry, Chiang Mai University, Chiang Mai, Thailand 50200, Thailand.
Corresponding author: Siriwoot Sookkhee, E-mail: siriwoot.s@cmu.ac.th
Total Article Views
Editor: Korakot Nganvongpanit,
Chiang Mai University, Thailand
Article history:
Received: June 9, 2021;
Revised: August 29, 2021;
Accepted: August 31, 2021;

