
Characterization of X- box binding protein 1(XBP1) and BiP functions and investigation their roles in YHV replication in P. monodon
Piyawadee Rugsanit, Pongsopee Attasart, Apinunt Udomkit, and Witoon Tirasophon*Published Date : 2021-09-13
DOI : https://doi.org/10.12982/CMUJNS.2021.090
Journal Issues : Number 4, October-December 2021
Abstract Eukaryotic cells have mechanisms to cope with stress in endoplasmic reticulum (ER) called unfolded protein response (UPR). In this study, we characterized putative X-box DNA binding protein1 (XBP1) and Binding protein (BiP) cDNAs from black tiger shrimp (Peneaus monodon). When the shrimp were infected with Yellow head virus (YHV), the levels of XBP1 and BiP mRNA transcripts were elevated approximately 8 and 55 folds, respectively. In normal shrimp, the putative XBP1 (XBP1u) was predicted to encode a protein with 283 amino acid residues. When shrimp were infected with YHV, portion of cDNA with 17 nucleotides intron elimination (XBP1s) was observed. The elimination caused alteration of its translational frame. The predicted protein from this is 469 amino acids in length with total change in its amino acid sequence downstream of the intron. Functional analysis of these XBP1 proteins in mammalian cells clearly showed that overexpression of P. monodon XBP1s was capable of activating the transcription rate of mammalian UPR responsive genes (BiP, ERdj4 and P58IPK). Finally, the impact of XBP1 and BiP on YHV multiplication in black tiger shrimp was investigated by RNAi approach. Knocking down either XBP1 or BiP expression efficiently inhibited YHV replication. Therefore, these two components in the UPR pathway may be an interesting target for anti YHV development in the future.
Keywords: Endoplasmic reticulum, siRNA, Stress-inducible genes, Transcriptional regulation
Citation: Rugsanit, P., Attasart, P., Udomkit, A., and Tirasophon, W. 2021. Characterization of X- box binding protein 1(XBP1) and BiP functions and investigation their roles in YHV replication in P. monodon. CMU J. Nat. Sci. 20(4): e2021090.
INTRODUCTION
An unfolded protein response (UPR) is a mechanism for maintenance homeostasis in endoplasmic reticulum (ER). In mammals, the UPR functions through three transmembrane sensors designated as protein kinase-like ER kinase (PERK), activating transcription factor 6 (ATF6) and inositol-requiring enzyme 1(IRE1) (Schröder et al., 2005; Walter et al., 2011). Amongst these, the cascade mediated through IRE1 is the most conserved branch found from yeast to mammal (Mori et al., 1993; Tirasophon et al., 1998; Plongthongkum et al., 2007). In certain condition when high levels of new proteins are synthesized and fluxed into ER, these proteins may not be properly folded. They retained in the organelle as unfolded/misfolded protein causing ER stress. Such condition deteriorates to cell survival and may lead to cell death (Urra et al., 2013). Several pathways of the UPR cooperatively function to overcome this threat by pausing further protein translation, eliminating unfolded protein and restoring the ER condition (Harding et al., 2005). Primarily, the IRE1 pathway upregulates the transcription of genes encoding ER-specific molecular chaperones and folding enzymes to match their requirements (Cox et al., 1996). The signature of the IRE1 is its endoribonuclease domain which initiates an unconventional splicing of intron in XBP1 mRNA to produce spliced XBP1mRNA (Kawahara et al., 1998). The spliced XBP1 mRNA is then translated into a potent transcription factor that upregulates transcription of ER chaperones and folding enzymes (Yoshida et al., 2001; Calfon et al., 2002; Lee et al., 2003).
Among conditions known to trigger ER stress, viral infections have been demonstrated to vigorously induce ER stress by forcing their host cells to produce high level of viral proteins for their propagation (Hurtley et al., 1989; Watowich et al., 1991; Kim et al., 2008). Several of the viral proteins must be translocated into ER for their folding (Inoue et al., 2013). Activation of the UPR pathway provides both pro and con to the viruses. Enhancement of the folding efficiency could benefit to viruses as the cell will be able to accommodate the viral proteins more efficiently. However, the activation usually accompanies with shutdown of new protein synthesis including the viral proteins to protect themselves from engaging in cell death pathway (Szegezdi et al., 2006. Nevertheless, infection with different types of viruses modulated the UPR pathway in diverse fashions (Estrabaud et al., 2011; Hassan et al., 2012; Hassan et al., 2014). For instance, Flavivirus activates XBP1 splicing (Yu et al., 2006). On the contrary, an infection with hepatitis C virus suppresses XBP1 splicing (Tardif et al., 2002), suggesting a complex relation between viral infections and XBP1. Because the RNase domain of IRE1 is similar to RNase L, which are involved in defense against viral infections, the UPR and antiviral system may share a common ancestral mechanism (Yoshida et al., 1998).
To date, the UPR pathway (IRE1/ XBP1 branch) and their impact on viral infection have been extensively characterized in mammalian cells, however, these information in other systems including crustacean were not well understood. Hence, it is not known whether this pathway in shrimp plays any role in viral infection. As mentioned above, distinct roles of UPR on different viruses were evidenced. In case that the pathway is crucial for YHV infection, understanding their relationship may benefit to the development of a new strategy to control this viral disease in black tiger shrimp in the future.
MATERIALS AND METHODS
Yellow head virus stock
YHV stock was prepared as previously described by Posiri et al. (2007). Briefly, YHV were collected from hemolymph of moribund shrimp infected with YHV. The virus was purified by Urogeafin® gradient ultracentrifugation. The YHV pellet was collected and resuspended in TNE buffer (10 mM Tris-HCl, pH 7.4, 100 mM NaCl and EDTA) and stored at -80oC. The YHV level in the stock (2 x 109 TCID50/ ml) was determined by Tissue culture infectious dose method (TCID50) in Penaeus monodon primary lymphoid cells. The purified virus was prepared as 10 folds serial dilution in the TNE buffer. The amount of virus that caused 100% mortality in shrimp within 4 days post injection was used in this study.
Shrimp rearing, gene knockdown by double-stranded RNA injection and YHV challenge
Healthy juvenile black tiger shrimp, P. monodon (size approximately 1 g) were reared in individual cages in plastic tanks. The tanks were filled with aerated artificial sea water at 10 ppt salinity. The animals were fed daily with commercial diet. Before each experiment, the animals were checked by PCR or RT-PCR method (24) to assure that they were not infected with YHV and white spot syndrome virus (WSSV). To infect the shrimp, the viral dose casing 100% mortality in 4 days was intramuscularly injected into the animals. The YHV level in gill of the infected shrimp were monitored by RT-PCR or qRT-PCR.
For RNAi experiment, shrimp were injected with indicated dsRNA at 2 μg/ shrimp (10 shrimp/group). At 48 hours post-injection, the shrimp were reinjected with the same of dsRNA containing YHV at the same dose as mentioned above. Gills from these shrimps were collected for further analysis. For mortality assay, after the second injection, viability of the shrimp was observed twice a day and monitored for 7 days.
RNA preparation, RT-PCR and qRT-PCR
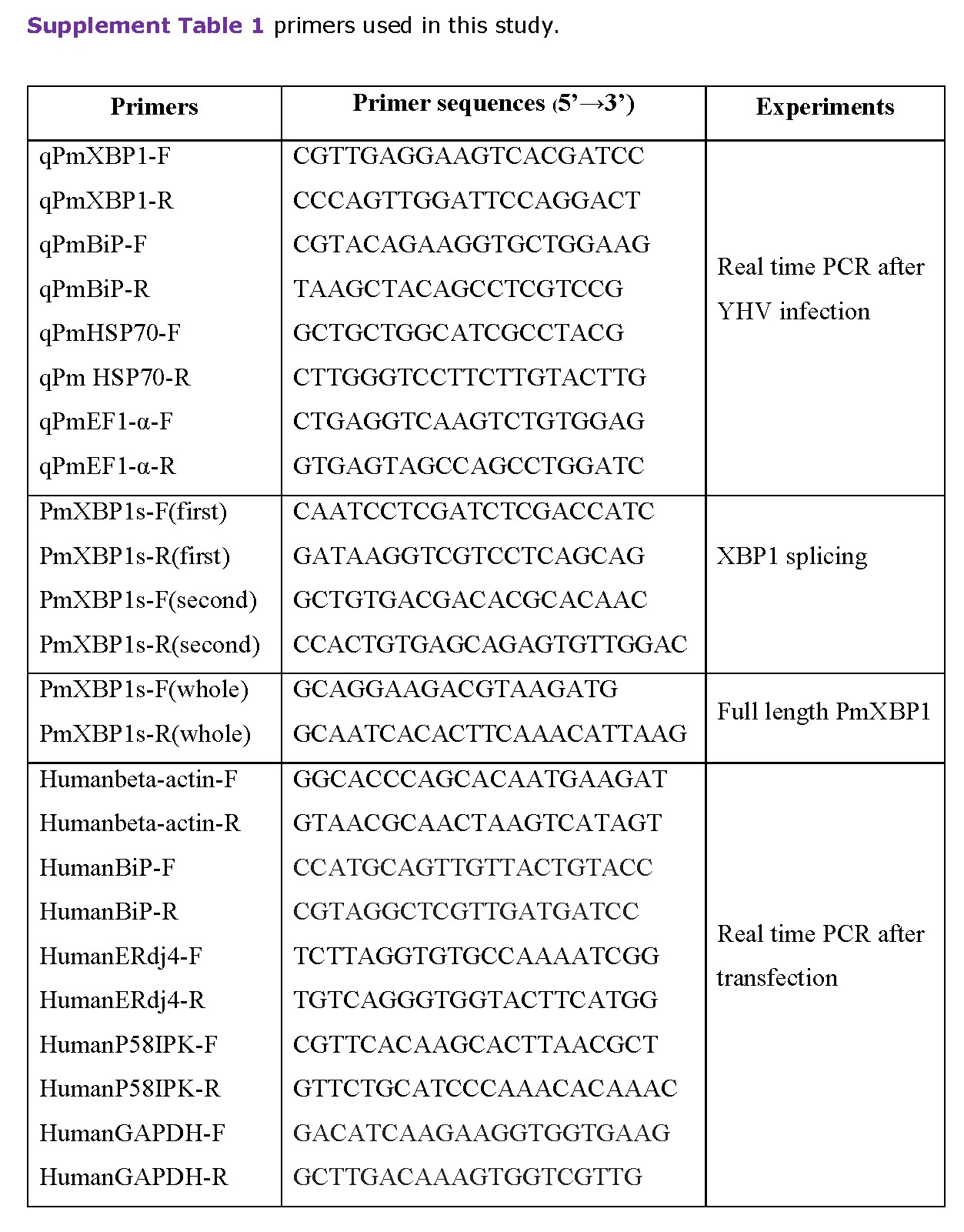
Total RNA was isolated from gill of individual shrimp using TRI-LS reagent (Molecular Research Center). cDNA was synthesized from one microgram of total RNA in a 20 μl reaction containing oligo-dT and Imprompt II reverse transcriptase (Promega). The cDNA sample was monitored by multiplex PCR using gene specific primers (supplement table 1). To monitor the presence of two forms of putative XBP1, 30 cycles of PCR amplification was performed. The PCR products were analyzed by 3% agarose gel electrophoresis.
qRT-PCR was performed using SYBR® Fast master mix qPCR kit (KAPA Biosystems) in 96-well plates. Each 20 μl PCR reaction contained 5 μl of diluted cDNA, 0.3 µM of forward and reverse primers and 10 μl of 2xSYBR Green master mix. The temperature profiles were started with denaturation at 95°C for 3 minutes. The reactions were then amplified for 40 cycles at 95°C for 10 seconds, 60°C for 30 seconds and 72°C for 20 seconds using Eppendorf Mastercycler® ep realplex. The cycle threshold (Ct) value was used to determine the expression level of the targeted gene. EF1-α level in each sample was an internal control. The alteration of the expression level was calculated by comparative method (2-ΔΔCT).
Plasmid construction
Recombinant plasmids expressing XBP1 was constructed in pcDNA3.1 (+) plasmid. The entire coding sequence of unspliced (or spliced) form of putative XBP1 was amplified from cDNA of YHV infected P. monodon. The entire sequence of each construct was verified by automated DNA sequencing.
Plasmid expressing dsRNA in E. coli was constructed in pET-17b vector (25). dsRNAs were expressed as hairpin RNA with approximately 200 bp stem and 100 bp loop (25-26). The recombinant plasmids were confirmed by DNA sequencing prior to transforming into the RNaseIII mutant E. coli HT115 strain. To induce the expression of the dsRNA, 0.1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) was added to the culture for 3 hours. The cell pellet was collected by centrifugation and used for RNA extraction by ethanol precipitation method. Host RNA in the sample was eliminated by addition of 0.005 μg RNaseA (Posiri et al., 2013).
Transient DNA transfection in COS-1 cells
COS-1, African green monkey kidney cells, were seeded in 6 well plate format. Upon the cell density reached 95% confluence, XBP1 expression plasmids were transfected into the cells by LipofectamineTM 2000 using protocol recommended by manufacturer (Poothong et al., 2017). At 48 hours after transfection, RNA were harvested from the transfected cells using TRIZOL reagent (MRC) (Poothong et al., 2017). The expression levels of some IRE1/XBP1 responsive genes (BiP, ERdj4 and P58IPK) in each condition were determined by qRT-PCR.
Statistical analysis
The relative mRNA levels of XBP1, BiP and gp116 normalized with PmEF1-α and fold change expressions were represented as mean ±SEM. Cumulative mortality were plotted as mean ±SEM. Significant differences between the data in each group of the experiment were tested by analysis of variance (ANOVA), and a probability (P) value of less than 0.05 was used to define significance.
RESULTS
YHV infection activates XBP1 and BiP expression in P. monodon
Putative XBP1 cDNAs showing low homology to Drosophila XBP1 were identified from EST library of P. monodon. Despite their overall homology were extremely low (5.9%), a highly conserved sequence of the deduced amino sequences (residues 90 to 160) was detected in theses cDNAs which are resembled to the basic leucine zipper region of XBP1s in other species (Figure 1). To investigate whether these cDNAs are XBP1 homolog, their ability to respond to ER stress were characterized. Previously, we demonstrated that shrimp infected with YHV produced high level of YHV structural proteins including GP116. This glycoprotein is a major component of YHV capsid which must be folded and modified with N-linked glycosylation in ER. Therefore, synthesis of these viral proteins at high level most likely induces ER stress in shrimp cells. Hence, YHV infection was used as condition the dissect to role of putative XBP1 in black tiger shrimps.
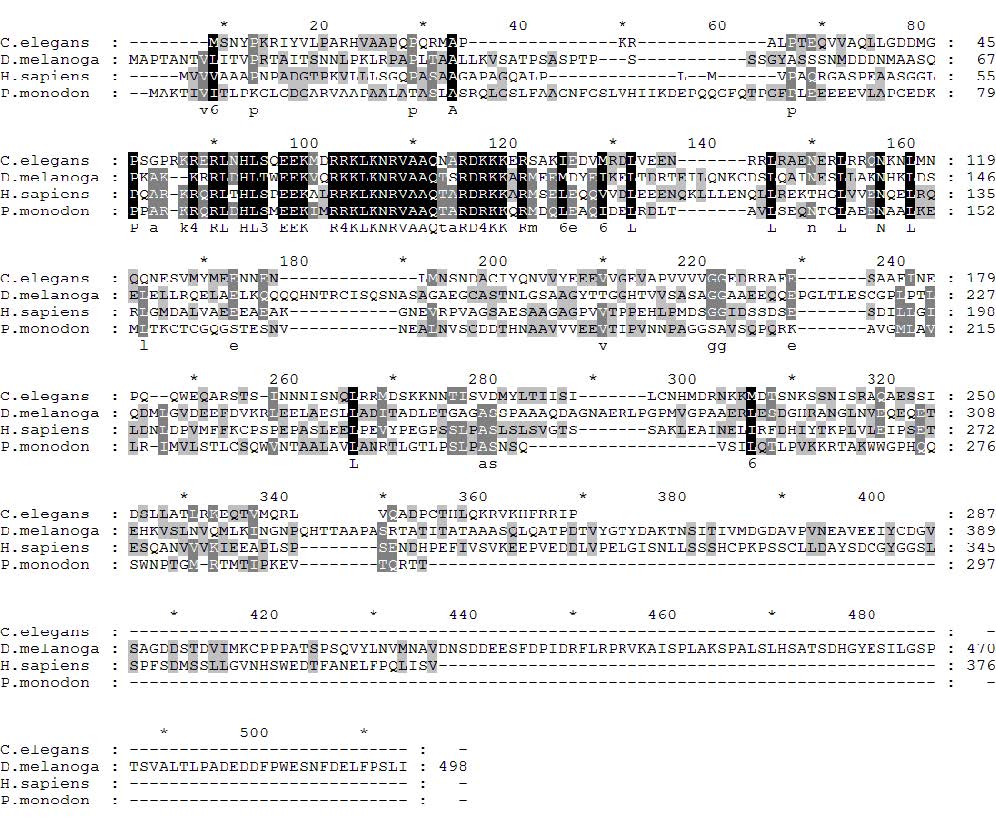
Figure 1. Alignment of deduced amino acid sequence of putative XBP1 from P. monodon and three well characterized XBP1 homologs (Human (Accession # NP_001073007.1), worm (Accession # CAL63999.1) and fruit fly (Accession # NP_726032.4). The highly conserved region among these proteins were presented in dark highlight.
The expression profiles of the putative XBP1 homolog from black tiger shrimp were monitored at different time intervals post-YHV infection. Upon infected with YHV, the level of the putative XBP1 mRNA in the shrimp was increased approximately 9 folds at 40 hr post infection (hpi) (Figure 2). The kinetic of this mRNA induction is resembled to that of BiP mRNA, a well characterized ER stress responsive gene. However, the transcriptional induction of BiP was detected at much higher level (60 folds). The transcriptional induction upon YHV infection appeared specific to certain genes particularly those encoding ER specific proteins. This condition had no effect to the expression level of HSP70, a BiP counterpart that specifically functions in cytosol. HSP70 mRNA level in normal shrimp appeared stable for the entire period of our study. Based on these evidences, it was clear that YHV infection triggered ER stress in black tiger shrimp, and the condition upregulated the expression of BiP and the putative XBP1.
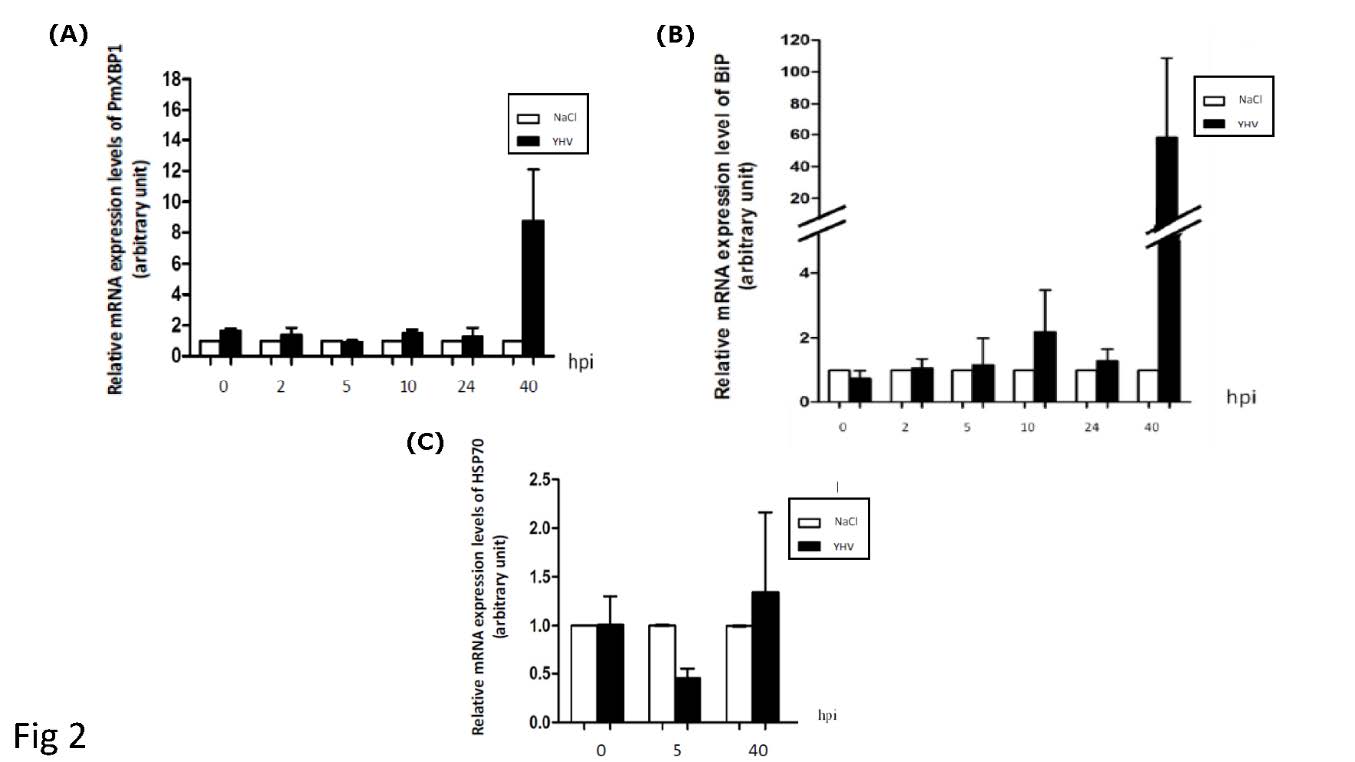
Figure 2. YHV infection in P. monodon activates of XBP1 and BiP expression. Relative mRNA level of XBP1 (A), BiP (B) and HSP70 (C) in gill of shrimp upon YHV infection were monitored post-YHV infection (hpi) by qRT-PCR. The expression level of each gene was normalized with that of EF1-α and presented as fold change comparing to the corresponding mRNA from shrimp injected with NaCl (NaCl). Average values were presented (n=10).
Besides transcriptional upregulation, the presence of two forms of putative XBP1 cDNA were detected in YHV infected shrimps (Figure 3A). A long form that was found in both normal shrimp and YHV uninfected shrimp. A short form was specifically detected in the infected shrimp (around 40 hpi) (Figure 3B). Sequence analysis revealed only 17 nucleotides difference between the two forms (Figure 3A and Supplement figure 1). The longer form of putative XBP1 contained an intact stretch of 17 nucleotide residues (between residue 602 to 618 relative to ATG translational start codon) within its ORF (Supplementary figure 1). The shorter form of putative XBP1, on the other hand, lacked these 17 nucleotide residues in its ORF. As a result, its deduced amino acid sequences downstream to this deletion was totally altered. Interestingly, these 17 nucleotide residues together with their flanking regions were predicted to form double hairpin structures with 7 nucleotides ring resembled to the spliced junction of well characterized XBP1 mRNAs from other organisms (Figure 3C). More importantly, the nucleotide residues within the 7 nucleotide rings which were previously characterized as conserved residues in other XBP1 mRNAs were absolutely conserved in the putative shrimp XBP1 mRNA as well. Taken together, it is convincingly that the two populations of putative XBP1 cDNA represent unspliced XBP1 (XBP1u) and spliced XBP1 (XBP1s) for P. monodon.
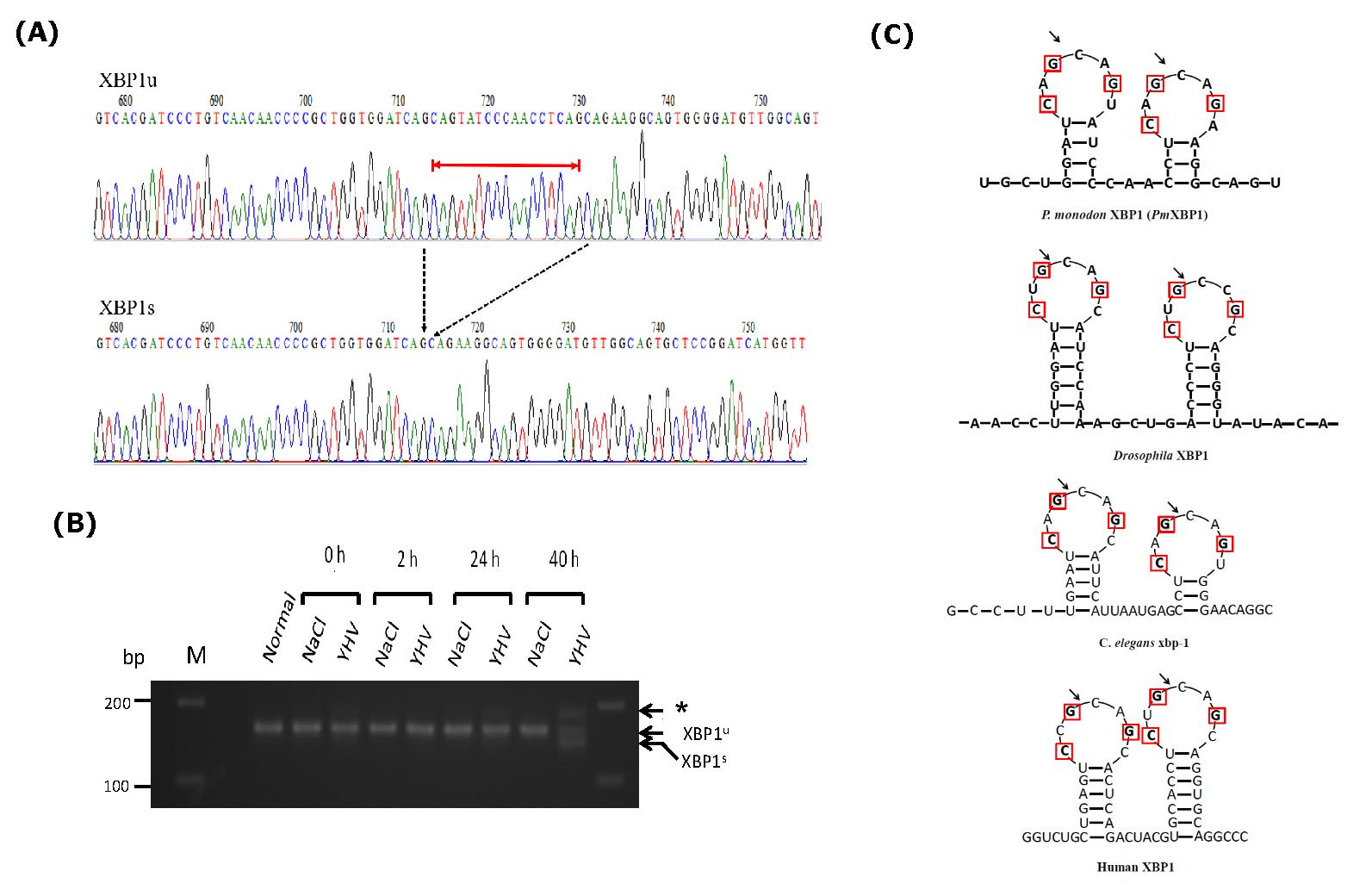
Figure 3. YHV infection activates IRE1/ XBP1 pathway in P. monodon. (A) Chromatograms showing nucleotide sequences from two populations of XBP1 cDNAs isolated from gill of YHV infected shrimp. The alignment showed only the region covering the different between the two cDNAs. The 17 nucleotides indicated with red line were absence in the short form (XBP1s)( Accession #MZ560705) compared to the long form (XBP1u)(Accession #MZ560704). Dash arrows indicated the position where the fragment was deleted from XBP1u. (B) Comparison of RT-PCR product of XBP1 from YHV infected shrimps (YHV) and control shrimp (NaCl) at different hours post infection. Deletion of 17 nucleotides in XBP1 mRNA were specially detected at late stage (40 hr.) of YHV infection.
* represents duplexes between XBP1u and XBP1s. (C) The predicted 7 membered ring secondary structures of XBP1u mRNAs from different organisms covering their unusual intron. Red boxes indicated the conserved residues found in known XBP1 mRNAs. Arrows indicated the potential spliced sites on XBP1 mRNA that were cleaved by IRE1 endoribonuclease.
P. monodon spliced XBP1 encodes an active transcription factor that activates transcription of UPR target genes
As mentioned earlier, the deletion of 17 nucleotides within the ORF of putative P. monodon XBP1mRNA caused translational frameshift. This led to alteration of the predicted size of protein product as well as their deduced amino acid sequences. XBP1u was predicted to encode only 283 amino acid residues whereas XBP1s was predicted to encode 469 amino acid residues (Figure 4A). The two proteins shared only 201 residues on their amino termini covering the highly conserved basic leucine zipper. It was interesting to further investigate the role of these 2 proteins. Unfortunately, appropriate cultured shrimp cells for their functional analysis are not existed. Therefore, functional analysis of these proteins was then investigated in mammalian cells instead. To precisely demonstrate the function of each shrimp XBP1 in UPR pathway, the intact coding sequences of the two cDNAs were cloned into pcDNA 3.1(+). The recombinant plasmids were transfected to overexpress their protein products in mammalian (COS-1) cells. In this study, the transcriptional activation properties of their protein products were analyzed from their ability to upregulate transcription of 3 well characterized mammalian UPR responsive genes (BiP, ERdj4 and P58IPK). The result in figure 4B showed that overexpression of the putative XBP1s alone significantly enhanced expression of these UPR responsive genes (between 2 to 6 folds). In contrast, overexpression of the putative XBP1u had no significant effect on transcriptional activation of these UPR target genes. To this point, it is quite certain that the two shrimp mRNA populations identified in our study are XBP1 homologous.
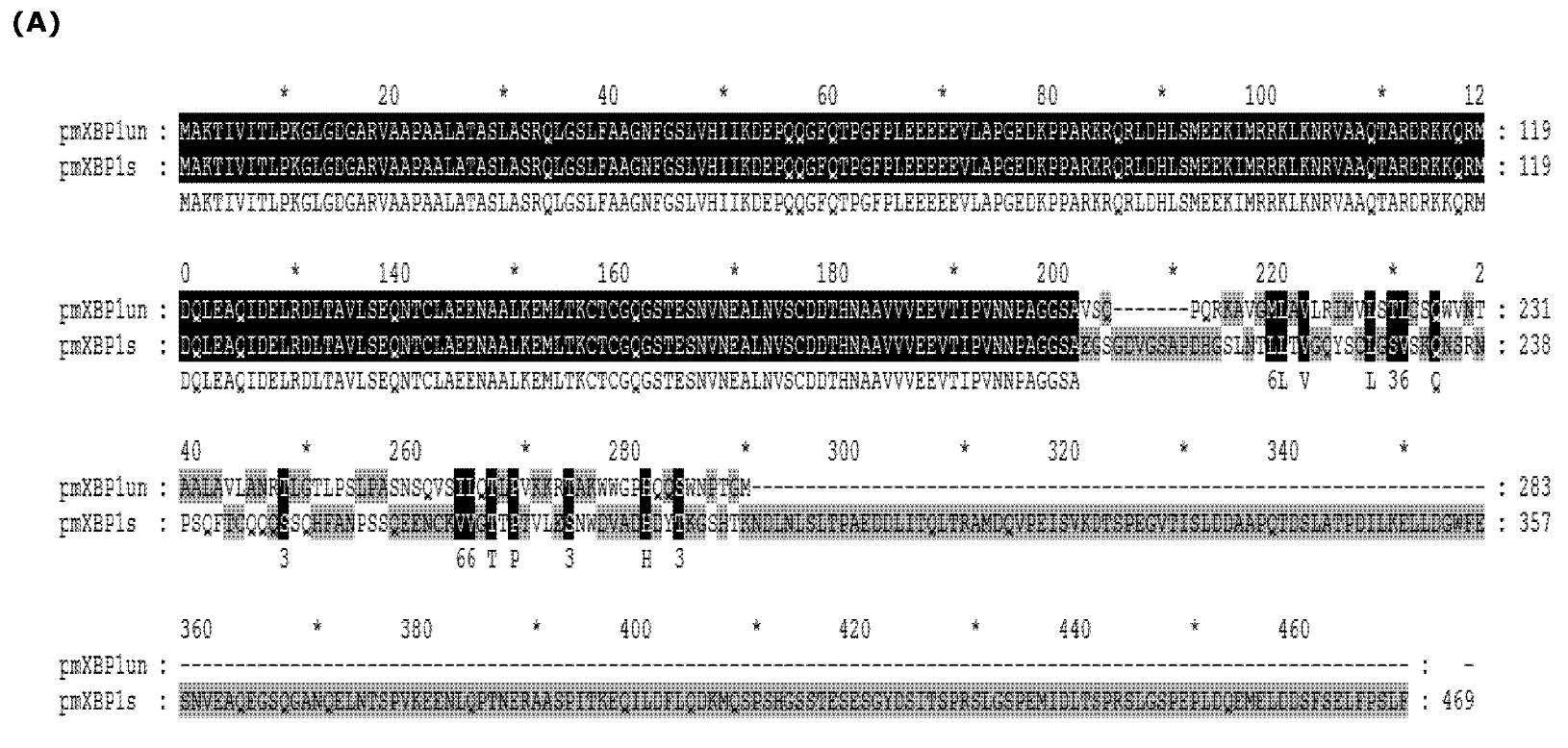
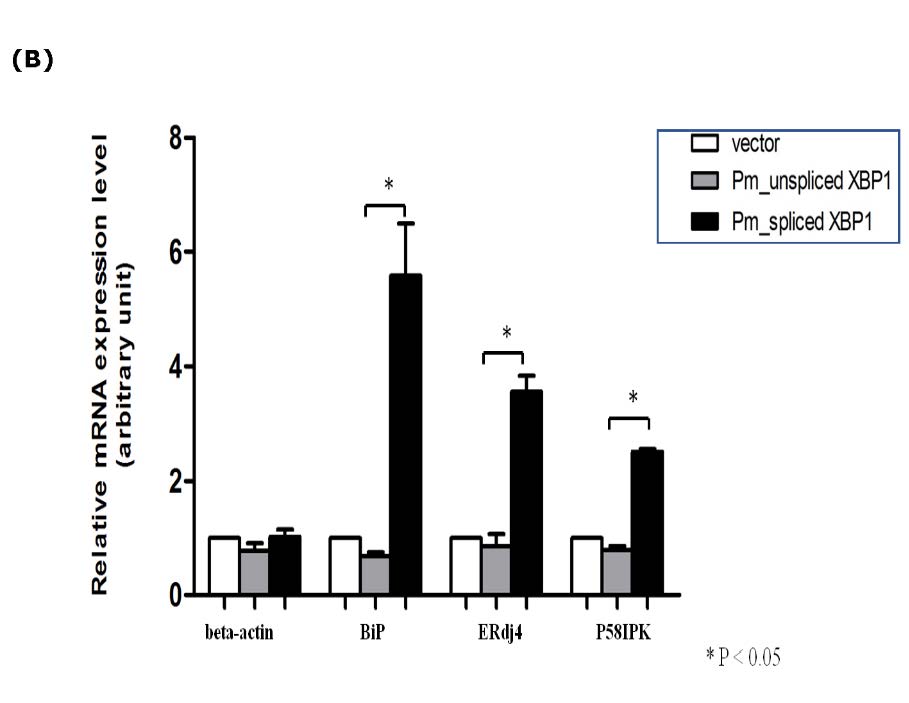
Figure 4. Overexpression of P. monodon XBP1s activated transcription of UPR target genes in COS1 cells. (A) Alignment of predicted amino acid sequences of spliced (XBP1s) and unspliced form (XBP1u). The XBP1u encoded a protein with 283 amino acid residues (indicated as pmXBP1un) while XBP1s encoded 469 amino acid residues (indicated as pmXBP1s). These two forms of XBP1 proteins shared 201 amino acid residues on their N termini as shown in dark highlight. (B) The entire coding sequences of XBP1u or XBP1s were cloned into mammalian expression plasmid and expressed in COS-1 cells by transient transfection. The transcriptional activation of 3 known targets for XBP1 transcription factor (BiP, ERdj4 and P58IPK) and actin were monitored by qRT-PCR. Relative expressions for each target gene were presented in average value with standard error from duplicated experiments. Significant change in the expression level was indicated by * (P value < 0.05).
The UPR pathway in black tiger shrimp cell is crucial for YHV replication
The finding of XBP1 and BiP transcriptional activation in shrimp during YHV infection suggested that the UPR may engage in YHV propagation. To decipher the role of these components in YHV infection, RNAi approach was used to suppress their expressions during the viral infection process. Initially, the RNAi efficiency was determined by injecting specific dsRNAs (dsRNA-XBP1, dsRNA-BiP or unrelated dsRNA-GFP) into black tiger shrimps (2 µg dsRNA/ g shrimp) (n =10 per group). As expected, the targeted mRNAs were dramatically reduced to their minimal levels (approximate 3% for XBP1 and 6 % for BiP compared to their normal levels) at 48 hours post-dsRNA injection (data not shown). The knockdown effect of each target was retained at least 72 and 96 hours for BiP and XBP1, respectively. However, injection with unrelated dsRNA-GFP had no effect on the expression level of either XBP1- or BiP mRNA indicating that the gene knockdown effects occurred in sequence specific manner. Therefore, dsRNA-GFP injection was used as a control group in all experiments.
Next, an impact of XBP1- or BiP knockdown on YHV propagation was analyzed in shrimp infected with this virus. Injection with unrelated dsRNA-GFP in the presence of YHV infection did not significantly affect to the expression level of either XBP1-or BiP mRNA. This result confirmed that suppression of XBP1 and BiP expression occurred in sequence specific fashion (Figure 5). YHV levels in these shrimp were comparable to that in the YHV infected shrimp without any dsRNA injection. In contrast, injection of either dsRNA-XBP1 or dsRNA-BiP prohibited transcriptional upregulation of their corresponding mRNAs in YHV infected shrimps. To this end, it was clear that suppression of either XBP1 or BiP expression inhibited YHV propagation in black tiger shrimp. While BiP knockdown showed complete inhibition of YHV propagation in shrimp, XBP1 knockdown only clearly reduced the viral level in the infected animals. Besides their ability to inhibit YHV replication, it was worth noting that dsRNA-XBP1 injection not only blocked its own mRNA induction in YHV infected shrimps but also prohibited BiP mRNA induction in infected shrimps. This result confirmed that P. monodon XBP1 is an upstream transcription factor that modulates transcriptional activation of BiP mRNA during ER stress.
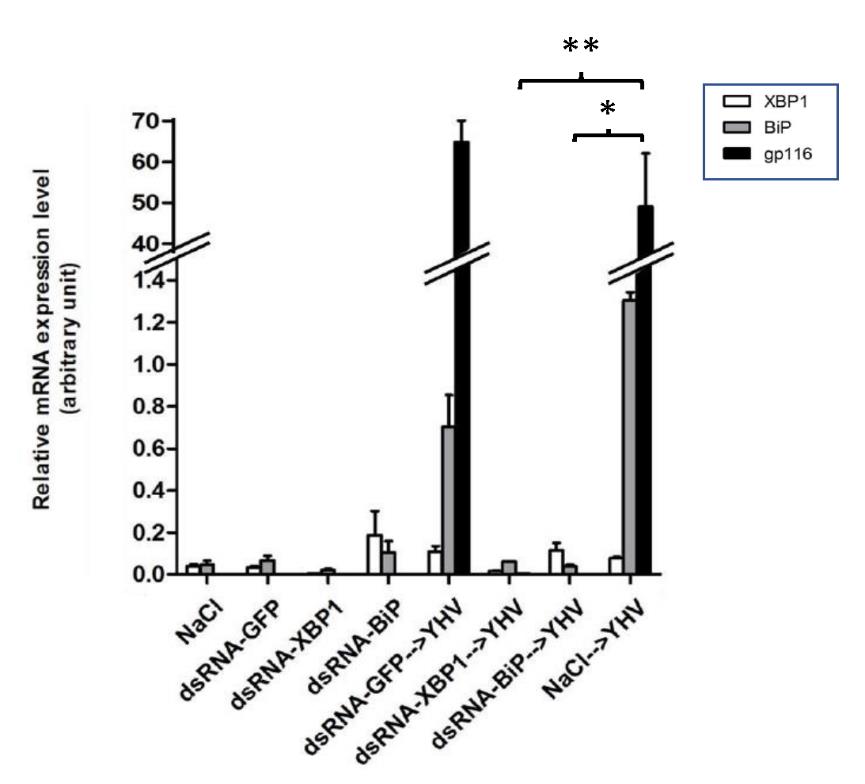
Figure 5. Suppression of XBP1 and BiP by RNAi prohibited YHV replication in black tiger shrimp. Shrimp (n=10) were injected twice with indicated dsRNA at 2 µg/g shrimp in 48 hr. interval. The second dsRNA injection was performed together with YHV challenge. The levels of XBP1, BiP and YHV-gp116 mRNAs in gill of all shrimp were determined by qRT-PCR and normalized with EF1-α. The data was shown in average value of duplicated experiments. * and ** represent P value
In addition to the analysis at the level of RNA transcript, mortality rate in the knock down animals (with or without YHV challenge) was also determined. Injection of either dsRNA-XBP1 or dsRNA-GFP alone caused no harm to the shrimps. All animals appeared strong and healthy until the end of the experiment (7 days). In contrast, injection with dsRNA-BiP resulted in 60 to 100% shrimp mortality (varied from experiment to experiment) (Figure 6B). The shrimp those still alive in this group were in moribund state (data not shown). This result suggested that BiP expression was extremely important to the shrimp therefore its dramatic reduction led to mortality. For YHV infected group, all shrimp with BiP knockdown died within 5 days after YHV challenge. In conjunction with the mentioned data, it was most likely that the mortality found in this group was resulted from BiP suppression not from the viral infection. In contrast, approximately 60% mortality in XBP1 knockdown or 100% in GFP knockdown shrimp were directly resulted from the viral infection (Figure 6A). Even though suppression of XBP1 did not result in absolute protection of shrimp mortality from YHV infection, a clear and consistent reduction in the mortality rate (~40%) suggested that XBP1 plays an important role in YHV propagation.
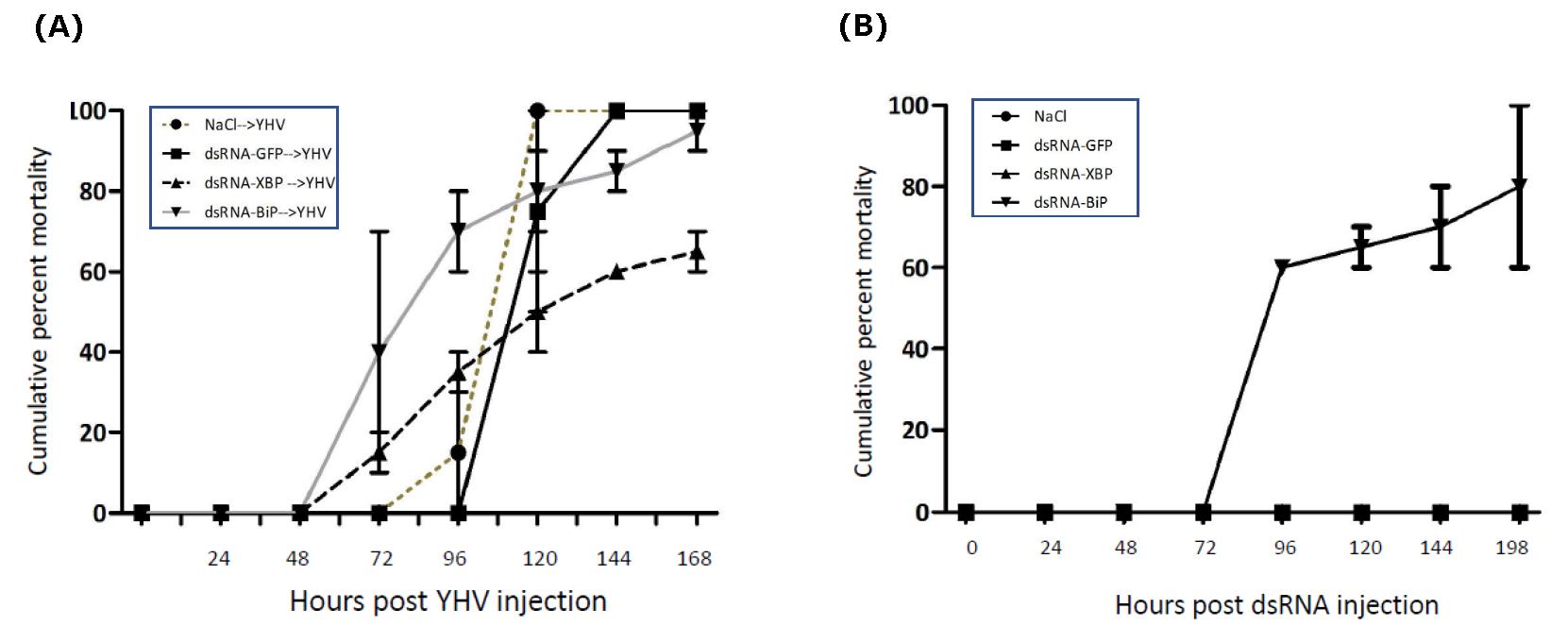
Figure 6. Mortality of XBP1 and Bip knocked-down shrimp upon YHV infection (A) and without YHV infection (B). Shrimp (n = 10 /group) were injected twice with indicated dsRNA at 2 µg/g shrimp in 48 hr. interval. The second injection was performed together with YHV challenge. Shrimp mortality was recorded twice a day for 7 days. Percent mortality in each experimental group was presented as means with error bar of duplicate experiments. Experiments in all groups were performed at the same time.
DISCUSSION
Transcriptional activation of genes in the UPR pathway is a key mechanism to adjust molecular chaperones and folding enzymes in ER to the levels which they are required. This mechanism is essential for survival of cells particularly in conditions promoting ER stress including viral infection. In black tiger shrimp, the role of UPR pathway in YHV infection particularly the cascade mediated through IRE1/XBP1 branch has not been previously investigated. In this study, we demonstrated that P. monodon XBP1, a bzip transcription factor, was responsible for transcriptional activation of ER specific molecular chaperone (BiP) during ER stress and XBP1 played crucial role in YHV replication process. The presence of spliced XBP1 specifically in YHV infected shrimp together with the functional analysis in COS-1 cells supported our hypothesis that YHV infection triggered the splicing of unorthodox 17 nucleotides intron in XBP1 mRNA through the IRE1/XBP1 cascade.
The result of XBP1- or BiP knockdown in YHV infected shrimp indicated that these two genes were vital to YHV propagation. Particularly, suppression of BiP expression completely abolished YHV multiplication. It was most likely that abrupt diminishing of this molecular chaperone paused overall protein folding in ER including several YHV proteins thus blocking the viral multiplication. It is generally accepted that BiP is an essential chaperone which participates in folding process of membranous and secretory proteins in ER (Munro et al., 1986). Therefore, its depletion by RNAi directly disturbed ER homeostasis which led to shrimp lethality (Li et al., 1991; Park et al., 2017; Kawaguchi et al., 2020). High mortality rate in BiP knockdown shrimp even without YHV infection in figure 6 strongly supported this notion. On the contrary, XBP1 knockdown caused no harm to the shrimp but clearly inhibited the viral propagation in the infected animal. Notably, we consistently found that XBP1 knockdown did not completely block YHV multiplication. Indeed, low level of the virus was detected in the knocked-down shrimp (data not shown). It was most likely that XBP1 knockdown did not pause all protein folding in ER. It only abrogated the ability of elevation of ER chaperones mediated through IRE1-XBP1 pathway during ER stress. Hence, the ER chaperones from their basal expression as well as from other pathways remained intact and functioned normally. From this scenario, these ER chaperones may be sufficient to accommodate ER protein folding as well as YHV propagation to some extent (Fros et al., 2015).
To this end, it is quite clear that IRE1/XBP1 pathway plays an important role in YHV propagation. However, additional experiments may be required to clarify how exactly this pathway engages in the viral propagation. First, the knockdown approach that is specifically target to only the spliced (or unspliced) XBP1 mRNA in the shrimp may be developed. Because the dsRNA induced XBP1 knockdown in our RNAi unable to selectively destroy each form of XBP1 mRNA. Even though there is no obvious evidence showing that protein translated from P. monodon unspliced XBP1 is involved in transcriptional activation especially during ER stress, however, this possibility cannot be totally excluded. Secondly, it was clear that the knockdown of endogenous gene target such as XBP1 inhibited YHV propagation in shrimp with lower efficiency when it was compared to the RNAi which was directly targeted to the viral genes (Yodmuang et al., 2006). This is a major drawback of RNAi that was designed to target the host genes such as XBP1. A possible explanation is that the expression level of these gene can resume to its normal level once the effect of dsRNA declined. In our study, when XBP1 knockdown was monitored for longer period, YHV propagation was detected at relatively high level in several shrimps (data not shown). At present, it was not clear if this was totally due to the recovery of XBP1 expression. Therefore, a knockdown platform that offer long lasting RNAi effect may help to clarify this issue. However, it is also possible that YHV may develop certain mechanism which allows the virus to propagate without the requirement of IRE1/XBP1 function. Indeed, similar mechanism has been reported in several viruses for mammalian cells (Su et al., 2002; Hassan et al., 2012; Fros et al., 2015). A more insightful information regarding YHV propagation will be useful to development of strategy for YHV control in the future.
ACKNOWLEDGMENTS
This work was supported by Mahidol university and the office of higher Education Commission to WT. PR was supported by the Office of the higher Education Commission and Mahidol University under the National Universities Initiative and Strategic Scholarships Fellowships Frontier Research Networks (Specific of Southern region).
AUTHOR CONTRIBUTIONS
Conceived and designed the experiment: WT PS, Performed the experiments: PR, Analyzed the data: WT PR. Contributed reagents/ materials/analysis tool: PA. Wrote the paper: WT AU PA PR.
REFERENCES
Assavalapsakul, W., Smith, D.R., Panyim, S. 2003. Propagation of infectious yellow head virus particles prior to cytopathic effect in primary lymphoid cell cultures of Penaeus monodon. Diseases of aquatic organisms. 55: 253–258.
Assavalapsakul, W., Smith, D.R., Panyim, S. 2006. Identification and Characterization of a Penaeus monodon Lymphoid Cell-Expressed Receptor for the Yellow Head Virus. Journal of virology. 80: 262–269.
Attasart P, Namramoon O, Kongphom U, Chimwai C, Panyim S. 2013. Ingestion of bacteria expressing dsRNA triggers specific RNA silencing in shrimp. Virus research.171: 252-256.
Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark, S. G., Ron, D. 2002. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 415: 92-96.
Cox JS, Walter P.1996. A Novel Mechanism for Regulating Activity of a Transcription Factor That Controls the Unfolded Protein Response. Cell. 87: 391-404.
Estrabaud E., De Muynck S., Asselah T. 2011. Activation of unfolded protein response and autophagy during HCV infection modulates innate immune response. Journal of hepatology. 55: 1150-1153.
Fros JJ, Major LD, Scholte FEM, Gardner J, van Hemert MJ, Suhrbier A, Pijlman, G. P. 2015. Chikungunya virus non-structural protein 2-mediated host shut-off disables the unfolded protein response. The Journal of general virology. 96: 580-589.
Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron, D. 2000. Regulated Translation Initiation Controls Stress-Induced Gene Expression in Mammalian Cells. Molecular cell. 6: 1099-108.
Hassan IH, Zhang MS, Powers LS, Shao JQ, Baltrusaitis J, Rutkowski DT, Legge, K., Monick, M. M. 2012. Influenza A viral replication is blocked by inhibition of the inositol-requiring enzyme 1 (IRE1) stress pathway.The Journal of biological chemistry. 287: 4679-4689.
Hassan I, Gaines KS, Hottel WJ, Wishy RM, Miller SE, Powers LS, Rutkowski, D. T. Monick, M. M. 2014. Inositol-requiring enzyme 1 inhibits respiratory syncytial virus replication. The Journal of biological chemistry.289: 7537-7546.
Hurtley SM, Bole DG, Hoover-Litty H, Helenius A., Copeland, C. S. 1989. Interactions of misfolded influenza virus hemagglutinin with binding protein (BiP). The Journal of cell biology. 108: 2117-2126.
Inoue T, Tsai B. 2013. How viruses use the endoplasmic reticulum for entry, replication, and assembly. Cold Spring Harbor perspectives in biology. 5: a013250.
Kawaguchi Y, Hagiwara D, Miyata T, Hodai Y, Kurimoto J, Takagi H, Suga, H., Kobayashi, T., Sugiyama, M., Onoue, T., Ito, Y., Iwama, S., Banno, R., Grinevich, V., Arima, H. 2020. Endoplasmic reticulum chaperone BiP/GRP78 knockdown leads to autophagy and cell death of arginine vasopressin neurons in mice. Scientific reports. 10: 19730.
Kawahara T, Yanagi H, Yura T, Mori K. 1998. Unconventional splicing of HAC1/ERN4 mRNA required for the unfolded protein response. Sequence-specific and non-sequential cleavage of the splice sites. The Journal of biological chemistry. 273: 1802-1807.
Kim I, Xu W, Reed JC. 2008. Cell death and endoplasmic reticulum stress: disease relevance and therapeutic opportunities. Nature reviews Drug discovery. 7: 1013-1030.
Lee AH, Iwakoshi NN., Glimcher LH. 2003. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Molecular and cellular biology. 23: 7448-7459.
Li XA, Lee AS. 1991. Competitive inhibition of a set of endoplasmic reticulum protein genes (GRP78, GRP94, and ERp72) retards cell growth and lowers viability after ionophore treatment. Molecular and cellular biology. 11: 3446-3453.
Mori K, Ma W, Gething MJ, Sambrook J. 1993. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus Cell. CellPress. 74: 743-756.
Munro S, Pelham HR. 1986. An Hsp70-like protein in the ER: identity with the 78 kd glucose-regulated protein and immunoglobulin heavy chain binding protein. Cell. 46: 291-300.
Park HJ, Park JY, Kim JW, Yang SG, Jung JM, Kim MJ, Park, J. J., Koo, D. B. 2017. Regulation of the Endoplasmic Reticulum Stress by BIP/GRP78 is involved in Meiotic Maturation of Porcine Oocytes In Vitro. Development & reproduction. 21: 407-415.
Plongthongkum N, Kullawong N, Panyim S, Tirasophon W. 2007. Ire1 regulated XBP1 mRNA splicing is essential for the unfolded protein response (UPR) in Drosophila melanogaster. Biochemical and biophysical research communications. 354: 789-794.
Poothong J, Sopha P, Kaufman RJ. 2017. IRE1α nucleotide sequence cleavage specificity in the unfolded protein response. FEBS Letters. 591: 406-414.
Posiri P, Ongvarrasopone C, Panyim S. 2013. A simple one-step method for producing dsRNA from E. coli to inhibit shrimp virus replication. Journal of virological methods. 188: 64-69.
Schröder M, Kaufman RJ. 2005. ER stress and the unfolded protein response. Mutation research. 569: 29-63.
Su HL, Liao CL, Lin YL. 2002. Japanese encephalitis virus infection initiates endoplasmic reticulum stress and an unfolded protein response. Journal of virology. 76: 4162-4171.
Szegezdi E, Logue SE, Gorman AM, Samali A. 2006. Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO reports. 7: 880-885.
Tardif KD, Mori K, Siddiqui A. 2002. Hepatitis C virus subgenomic replicons induce endoplasmic reticulum stress activating an intracellular signaling pathway. Journal of virology. 76: 7453-7459.
Tirasophon W, Welihinda AA, Kaufman RJ. 1998. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes & development.12: 1812-1824.
Urra H, Dufey E, Lisbona F, Rojas-Rivera D, Hetz C. 2013. When ER stress reaches a dead end. Biochimica et Biophysica Acta. 1833: 3507-3517
Walter P, Ron D. 2011. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 334: 1081-1086.
Watowich SS, Morimoto RI, Lamb RA. 1991. Flux of the paramyxovirus hemagglutinin-neuraminidase glycoprotein through the endoplasmic reticulum activates transcription of the GRP78-BiP gene. Journal of virology. 65: 3590-3597.
Yodmuang S, Tirasophon W, Roshorm Y, Chinnirunvong W, Panyim S. 2006. YHV-protease dsRNA inhibits YHV replication in Penaeus monodon and prevents mortality. Biochemical and biophysical research communications. 341: 351-356.
Yoshida H, Haze K, Yanagi H, Yura T, Mori K. 1998. Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. The Journal of biological chemistry. 273: 33741-33749.
Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. 2001. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell.107: 881-891.
Yu CY, Hsu YW, Liao CL, Lin YL. 2006. Flavivirus infection activates the XBP1 pathway of the unfolded protein response to cope with endoplasmic reticulum stress. Journal of virology. 80: 11868-11880.
OPEN access freely available online
Chiang Mai University Journal of Natural Sciences [ISSN 16851994]
Chiang Mai University, Thailand
Supplement Figure 1. An alignment of nucleotide sequences of spliced and unspliced XBP1 cDNA from black tiger shrimp (Penaeus monodon). 17 nucleotides intron corresponding to position +705 to +722.
Supplement figure 2. Deduced amino acid sequence of spliced XBP1 from black tiger shrimp.

Piyawadee Rugsanit, Pongsopee Attasart, Apinunt Udomkit, and Witoon Tirasophon*
Institute of Molecular Biosciences, Mahidol University, Nakhon Pathom, 73170 Thailand
Corresponding author: Witoon Tirasophon, E-mail: witoon.mu@gmail.com
Total Article Views
Editor: Korakot Nganvongpanit,
Chiang Mai University, Thailand
Article history:
Received: January 30, 2021;
Revised: August 11, 2021;
Accepted: August 16, 2021;

