
Toxicity and Anti-Oxidation Capacity of The Extracts from Caulerpa lentillifera
Supawadee Osotprasit, Tepparit Samrit, Athit Chaiwichien, Narin Changklungmoa, Krai Meemon, Nakorn Niamnont, Preeyanuch Manohong, Kunwadee Noonong, Montakan Tamtin, Prasert Sobhon, and Pornanan Kueakhai*Published Date : 2021-04-01
DOI : https://doi.org/10.12982/CMUJNS.2021.065
Journal Issues : Number 3, July-September 2021
Abstract Caulerpa lentillifera (sea grape) has been widely used in pharmaceutical industry and health-care products in Thailand. In this study, we attempted to evaluate the toxicity and antioxidant capacity of sea grape extracts in five fractions (ethanol- CLET, hexane- CLHE, ethyl acetate- CLEA, butanol-CLBU, and aqueous-CLAQ). The extracts were evaluated for cytotoxicity by MTT and LDH assays on four cell lines, fibroblast (L929), macrophages (RAW 264.7), hepatocytes (FL83B), and keratinocytes (HaCaT). Genotoxicity was tested by comet assay and micronucleus assay on human lymphoblast cells (TK6). The antioxidant capacity was measured by DPPH and ABTS scavenging assays. Our results demonstrated low cytotoxicity and genotoxicity of CLET, CLBU and CLAQ. When tested by DPPH and ABTS assays, CLET, CLEA, and CLHE showed high antioxidant activity. In conclusion, CLET, CLBU, and CLAQ demonstrated no toxic effects, and CLET, CLEA, and CLHE exhibited high antioxidant capacity. Therefore, our results indicated that CLET, CLEA, and CLHE could be consumed safely at doses lower than 500 and 200 μg/ml for CLHE and CLEA, respectively.
Keywords: Anti-oxidation, Caulerpa lentillifera, Cytotoxicity, Genotoxicity
Funding: This research was supported by the grants from the Thailand Research Fund (TRF)- Research and Researcher for Industry (RRi; MSD61I0029) and the Agricultural Research Development Agency (Public Organization).
Citation: Osotprasit, S., Samrit, T., Chaiwichien, A., Changklungmoa, N., Meemon, K., Niamnont, N., Manohong, P., Noonong, K., Tamtin, M., Sobhon, P., et al. 2021. Toxicity and anti-oxidation capacity of the extracts from Caulerpa lentillifera. CMUJ. Nat. Sci. 20(3): e2021065
INTRODUCTION
Marin algae or seaweeds are important parts of the ecological system. Seaweeds have been used commercially because they grow rapidly with high yield. In 2018, the export value of seaweed products worldwide was about USD 13.3 billion. From 2000 to 2018, the output of seaweeds has more than tripled, up from 10.6 million tonnes in 2000 to 32.4 million tonnes in 2018 (FAO, 2020). At present, many products are derived from seaweed, such as food, nutraceuticals, fertilizers, pharmaceuticals, cosmetics, and biochemicals. Seaweeds are also used for waste water treatment. Caulerpa lentillifera (sea grape) is a seaweed that can be found in coastal area of Thailand and has been cultured by Phetchaburi Coastal Fisheries Research and Development Center, Department of Fisheries and farmers since 1993. C. lentillifera is a popular edible species with high minerals, dietary fibers, vitamin A, vitamin C, and several essential unsaturated fatty acids (Matanjun et al., 2008). Previously, several biological activities of C. lentillifera extracts have been reported, including anti-cancer (Maeda et al., 2012), anti-oxidative (Matanjun et al., 2008), anti-diabetes (Sharma and Rhyu, 2014; Sharma et al., 2015; Nguyen et al., 2011), anti-coagulant (Arenajo et al., 2017), immuno- stimulatory (Sun et al., 2018; Maeda et al., 2012), and lipid-lowering activities (AbouZid et al., 2014; Matanjun et al., 2008; Nguyen et al., 2011; Sharma et al. 2015, 2017; Sharma and Rhyu, 2014). These biological activities suggest that C. lentillifera has high commercial values which can benefit farmers who grow these seaweeds in Thailand and beyond.
Toxicity or safety tests of raw materials before being processed into food and drug, and for environment safety are necessary. Especially for food and drugs, the selected extracts must not be cytotoxic and genotoxic and cause cell death and cell mutation. Previously, toxicity and genotoxicity were tested in laboratory animals. Because of ethical consideration on the use of animals, the cost and inconvenience wide range of in vitro tests have been developed and gain full acceptance to replace in vivo tests using animals (Walum et al., 1994; Collins et al., 2008).
Test for antioxidative capacity is also an important requirement for the extracts. Free radicals are atoms or molecules that have one or more unpaired electrons that can react quickly with other micromolecules and macromolecules. Free radicals are the by-products of metabolism and a harmful environment. It caused a lot of problems in an organism, such as DNA damage, mitochondria impairment, and misfolded proteins, induction of tissue injuries and many diseases by its activation of chronic inflammation (Arulselvan et al., 2016). Antioxidants are substances that can take up free radicals which are found in many plants, including C. lentillifera (Apostolova and Victor, 2015; Palanisamy et al., 2017). They can prevent and reduce many health problems generated by free radicals and inflammation.
Therefore, in this study, we aim to screen the toxicities and antioxidant capacity of solvent extracts from C. lentillifera. No research has been conducted to test each fraction of the extract toxicity and antioxidant. These fractions could be used as safe dietary supplements and raw materials for drug development for human and animal consumptions.
MATERIALS AND MENTHODS
Plant materials and extraction
C. lentillifera was pond-cultured by Ms. Montakan Tamtin, Phetchaburi Coastal Fisheries Research and Development Center, Thailand. The algae were extensively washed with tap water and dried at room temperature (RT). The dried C. lentillifera (1.00 kg) was air-dried, milled and macerated successively with 95% EtOH (3 L) at RT for 7 days. Next, EtOH extract was successively partitioned in solvents with increasing polarity from n-hexane, ethyl acetate, n-butanol and distilled water. The yields after evaporation of the solvents under reduced pressure were compared with dried weight of the seaweed, which for CLET (ethanol), CLHE (hexane), CLEA (ethyl acetate), CLBU (butanol), CLAQ (aqueous) were 12, 1, 0.5, 6.4, and 3.2%, respectively. The extracts were then filtered and evaporated under vacuum. Powders from the extracts were dissolved in 100% DMSO at 1 mg/ml and kept as stock solutions and stored at -20˚C.
Cell cultures
Mouse fibroblast (L929), mouse macrophages (RAW 264.7), mouse hepatocyte (FL83B), keratinocytes (HaCaT), and human lymphoblast cells (TK6) were purchased from American Type Culture Collection (ATCC). Briefly, L929, FL83B, and HaCaT cells were grown in DMEM culture medium with 1 g/L D-glucose, L-glutamine, 110 mg/L sodium pyruvate, penicillin G (10 U/ml), streptomycin (10 µg/ml), and 10% fetal bovine serum. RAW 264.7 and TK6 cells were grown in RPMI 1640 medium with L-glutamine, penicillin G (10 U/ml), streptomycin (10 µg/ml), and 10% fetal bovine serum. All cell lines were cultured at 37˚C in a humidified incubator with 5% CO2 atmosphere. The culture media were changed every 2–3 days. The cells at logarithmic phase were harvested immediately before performing each experiment.
MTT assay
The cell viability was measured by employing MTT assay using L929, RAW 264.7, FL83B and HaCaT cells. The cells were seeded into 96-well plates at 8X103 cells per well, and allowed to grow for 24 h at 37°C. Then, the medium was discarded, and cells were treated for 24 h with various concentrations of the extracts at 10, 50, 100, 200, 500, and 1,000 µg/ml in the culture medium, and 1% DMSO diluted in the culture medium as a negative control. MTT salt was dissolved in PBS (stock solution of 5 mg/ml). After treatments, the reaction medium was removed, the MTT was dissolved in culture medium (working solution: 10:100 μl v/v; 0.5 mg/ml), added to each well and incubated for 3 h at 37°C. Finally, the MTT was removed, and DMSO was added to solubilize formazan crystals. The plates were gently shaken for at least 15 min, and the OD were then read with a microplate spectrophotometer (VersaMax microplate reader) at a wavelength of 690 nm (background signal) and 570 nm. The percentage of viable cells was calculated after normalization with the negative control (1% DMSO), which was considered to have 100% cell viability.
LDH release assay
Cytotoxicity was analyzed by LDH assay (Sigma-Aldrich, Roche). L929, RAW 264.7, FL83B and HaCaT cells were seeded into 96-well plates at 8X103 cells per well in triplicate wells and allowed to grow for 24 h. Then, the cells were treated with various concentrations of the extracts with 10, 50, 100, 200, 500, and 1,000 µg/ml in the culture medium for 24 h. For controls the cells were incubated in 5 µl/well lysis buffer (positive control), 50 µl culture medium plus 50 µl assay medium (negative control) and incubated the plate for 15 min. A 50 μl of culture medium (sample and control) from each well was collected and transferred to a new 96-well plate. Then, 100 µl of the reaction mixture (freshly prepared) was added to each well on the 96-well plate and incubated for 5–10 min in a dark room. Finally, 50 µl stop solution was added to each well of the 96-well plate. Then, the OD were read by ELISA reader at 492 nm, and percent LDH was calculated for each sample.
Comet assay
Comet assay was carried out to measure the DNA damage. TK6 cells were seeded at 2×105 cells/ml in 24-well plate, the plate was maintained in a humidified incubator with 5% CO2 and 37°C for 24 hand treated with extracts at concentrations 10, 50, 100, 200, 500, and 1,000 µg/ml in the culture medium, hydrogen peroxide- H2O2 10 μM (positive control) and 10% FBS in RPMI medium (negative control). After incubation, the cells were collected, 100 μl of TK6 cell was centrifuged 4,500 rpm for 5 min, and the supernatant was discarded, and cells resuspend with 100 μl PBS pH 7.4. The cell suspension was mixed with 1% low melting point (LMP) agarose at 1:10 ratio at 37°C. And the same volume was deposited on a previously prepared thin layer 1% normal melting point (NMP) agarose, and then covered with a cover glass. The cover glasses were removed, and the slides were immersed in a freshly prepared lysis solution (2.5 M NaCl, 100 mM EDTA, 10 mM Tris (pH 10), supplemented with 1% Triton X-100) for overnight, at 4°C in the dark. The slides were then transferred to the unwinding solution (300 mM NaOH, 1 mM EDTA, pH > 13) which detected single and double stranded breaks, for 30 min at RT in a dark room. Then, electrophoresis was conducted under standard conditions (20 V; 300 mA; 1 Vcm−1) for 30 min. The slides were neutralized with 0.4 M Tris pH 7.5 for 5 min to remove alkali condition and detergents and stained with SYBR® Green, in the dark. The samples were examined with a fluorescence microscope (Olympus with 40X magnification) immediately, and analyzed with the Comet Score 2.0 software for determining DNA damage. At least 50 randomly non- overlapping cells per culture were examined in double blind observations. These cells were scored based on tail and head (nucleus) sizes. Following parameters were recorded: tail length, % of tail DNA, and tail moment, intensity of the fluorescencing DNA. Tail length was the length of the comet tail which indicated the extent of DNA damage. Percent of tail DNA was the intensity of fluorescing caused by damaged DNA in the comet tail, and the tail moment was calculated as (tail length) x (% tail DNA).
Micronucleus test (MN)
TK6 cells were seeded at 2X105 cell per ml in 24-well plate, then the plate was maintained in a humidified incubator with 5% CO2 at 37°C for 24 h. And then, the cells were treated with the extracts at concentrations 10, 50, 100, 200, 500, and 1,000 µg/ml in the culture medium, with 10 µg/ml mitomycin for positive control and 1% DMSO for negative control. After incubation, 6 µg/ml cytochalasin B was added into each well to obtain dividing binucleated cells for a period of 19 h. The cells were collected and 500 μl of TK6 cells were centrifuged at 4,500 rpm for 5 min, and the supernatant was discarded, cells resuspend in 500 μl PBS pH 7.4, and then centrifuged at 4,500 rpm for 5 min. A 500 μl of hypotonic solution (0.075M KCl) was added into each pellet and incubated at 37°C for 30 min and centrifuge at 4,500 rpm for 5 min. Next, the pelleted cells were fixed in methanol and acetic acid solution with the ratio 3:1. Finally, the slides were prepared by cytospin, stained with 10% Giemsa, and immediately examined in a light microscope (Olympus with 40X magnification). At least 50 cells were counted for each experimental and control sample, and the numbers of binucleated cells with micronucleus were recorded.
DPPH assay for antioxidant activity
Four sets of experiments were set up: I) five fractions of C. lentillifera extracts were diluted with methanol at 250, 500, 750, 1,000, 1,500, and 2,000 μg/ml. About 0.1 ml of each concentration of each extract was mixed with 0.1 ml of methanol in 96- well plate (designated as Sample+Met); II) A 0.2 ml of methanol was added in 96-well plate (designated as Met); III) 0.1 ml of DPPH working solution was mixed with 0.1 ml of methanol in 96-well plate (designated as DPPH+Met); and IV) each concentration of the extracts as mentioned in I and each concentration of positive controls ,vitamin C and quercetin, at concentrations 2, 4, 6, 8, and 10 μg/ml were diluted with 0.1 ml methanol, then they were mixed with 0.1 ml of DPPH working solution (designated as Sample+DPPH). After that, all samples were mixed and let stand at room temperature in the dark for 30 min. Finally, the absorbance of each group was measured at 517 nm. The EC50 value of each fraction was extrapolated from the linear equation by substituting the percent of scavenging effect (y) at 50%.
ABTS assay for antioxidant activity
Similar to DPPH assay, four similar sets of experiments were set up. Instead at the final step each sample was mixed with 0.1 ml of ABTS solution, and samples in the four sets were designated as Sample+Met, Met, ABTS+Met, Sample+ABTS. The samples were mixed and incubated at room temperature in the dark for 20 min, then the absorbance was measured at 734 nm. The EC50 value of each fraction was extrapolated from the linear equation by substituting the percent of scavenging effect (y) at 50%.
Statistical analysis
All data were presented as mean ± standard deviation (SD) and analyzed by GraphPad Prism 7 software (version 7.0). Comparisons between the extracts and controls were conducted using a one-way analysis of variance (ANOVA) followed by Duncan’s post hoc comparisons. Differences were considered statistically significant at P < 0.001.
RESULTS
Cytotoxicity of the extracts from C. lentillifera by MTT and LDH assays.
Cytotoxicity from C. lentillifera extracts at 24 h as measured by MTT assays (Figure 1) showed that for all cell types, IC50 of CLET, CLBU, and CLAQ were more than 1,000 µg/ml, when compared with the negative control. For L929 cells, IC50 of CLHE and CLEA were at 575 and 535 µg/ml, respectively. For FL83B cells, IC50 of CLHE and CLEA were at 514 and 250 µg/ml, respectively. For RAW 264.7 cells, IC50 of CLHE and CLEA were at 761 and 195 µg/ml, respectively. For HaCaT cells, IC50 of CLHE and CLEA were at 488 and 246 µg/ml, respectively. These results indicated that CLET, CLBU, and CLAQ were not cytotoxicity, while CLHE and CLEA have cytotoxicity at concentrations higher than 200 µg/ml.
LDH assay (Figure 2) showed that for all cell types, IC50 of CLET, CLBU and, CLAQ were more than 1,000 µg/ml when compared with the negative control. As well, for L929 cells, IC50 of CLHE and CLEA were at 566.3 and 435 µg/ml, respectively. For FL83B cells, IC50 of CLHE and CLEA were at showed as 995.4 and 629.6 µg/ml, respectively. For RAW 264.7 cells, IC50 of CLHE and CLEA were at 1,025 and 245.6 µg/ml, respectively. For HaCaT cells, IC50 of CLHE and CLEA were at 544.5 and 390 µg/ml, respectively. These results indicated that CLET, CLBU, and CLAQ were not cytotoxic, while CLHE and CLEA have cytotoxicity at concentrations higher than 200 µg/ml.
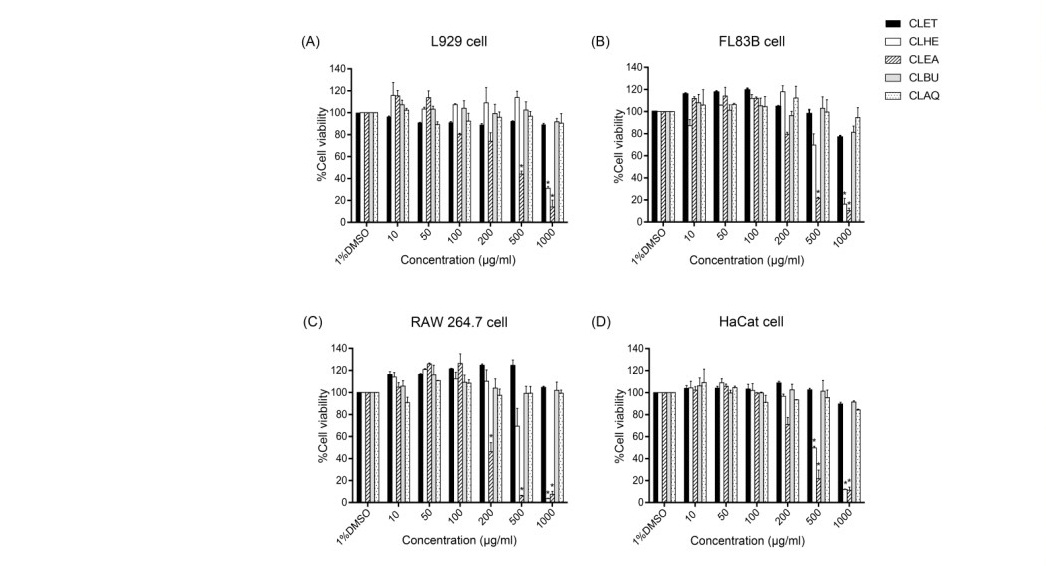
Figure 1. Cell viability of four cell types treated with C. lentillifera extract for 24 h, assessed by the MTT assay. (A) L929 cell line; (B) FL83B cell line; (C) RAW 264.7 cell line; (D) HaCaT cell line. Data was expressed as mean ± standard deviation of the percentage of cell viability in relation to 1%DMSO (100% cell viability),*P < 0.001
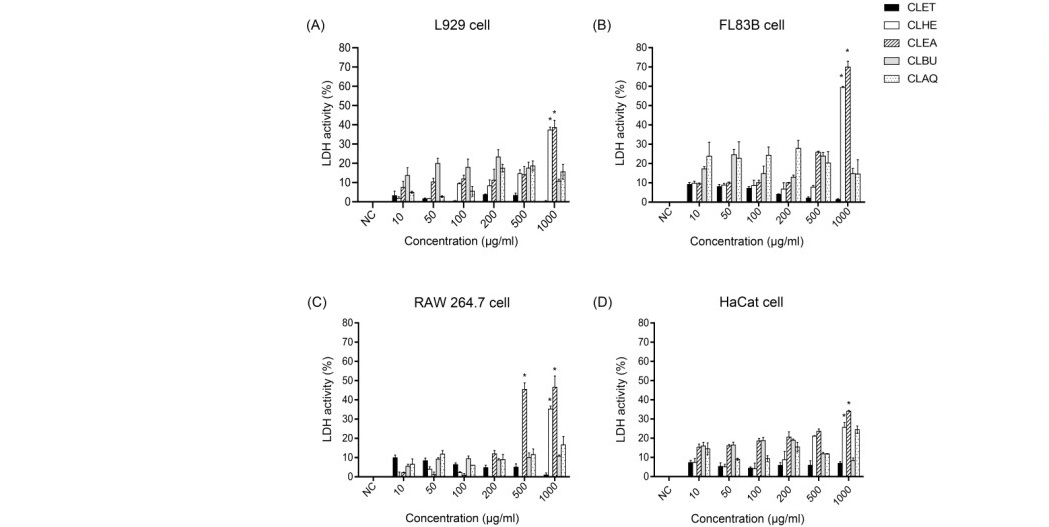
Figure 2. LDH activity of four cell types treated with C. lentillifera extracts for 24 h, assessed by the LDH assay. (A) L929 cell line; (B) FL83B cell line; (C) RAW 264.7 cell line; (D) HaCaT cell line. Data were expressed as mean ± standard deviation of the percentage of LDH activity in relation to the positive control (100% LDH activity), *P < 0.001
Genotoxicity of extracts from C. lentillifera estimated by comet assay
To evaluate whether the C. lentillifera extracts affected genes comet assay was performed with TK6 cell (Table 1). After treatment, three parameters were recorded, the most important of which was % tail DNA. Genotoxicities of CLET, CLBU, and CLAQ was slightly detectable at concentrations of 1,000 µg/ml, which showed comet score of less than 50 percent. For CLHE genotoxicity was detected at concentration of 500 and 1,000 µg/ml. For CLEA genotoxicity was detectable at concentrations of 200, 500, and 1,000 µg/ml. Both fractions showed comet score at more than 50 percent. These results indicated that CLET, CLBU, and CLAQ practically have no genotoxicity, while CLHE and CLEA were safe at concentrations less than 200 µg/ml.
Table 1. Genotoxicity measured by comet assay for tail length, % tail DNA and tail moment in TK6 cells treated with different concentration of C. lentillifera extracts for 24 h.
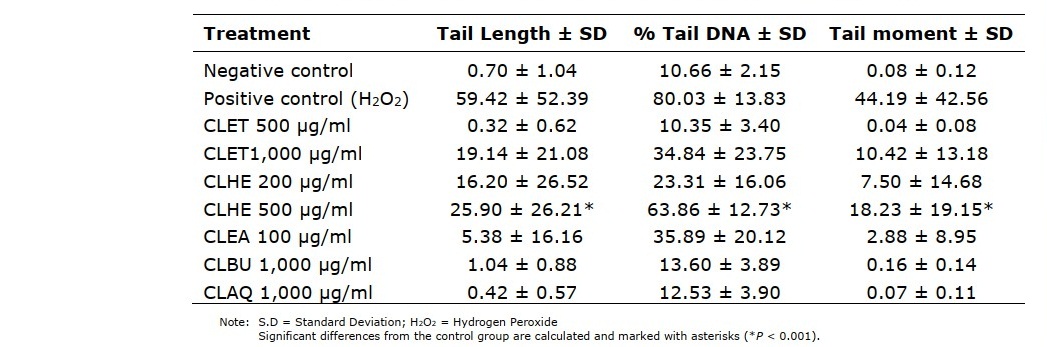
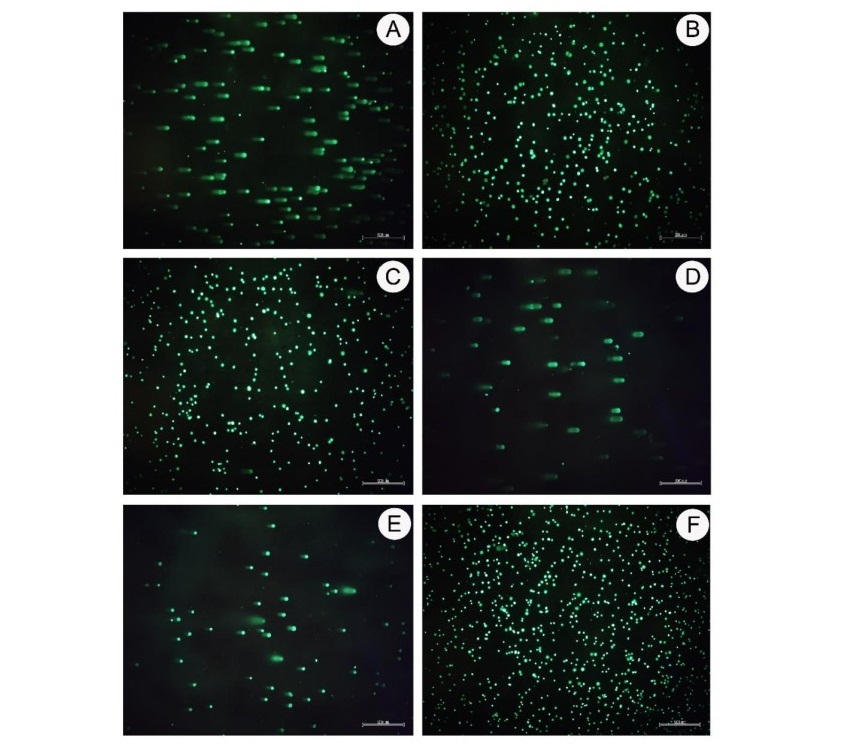
Figure 3. Genotoxicity measured by comet assay was performed with the TK6 cell. The DNA damage is expressed in a comet pattern due to strand breaking and alkylating agents as analyzed by Comet Score 2.0 software. (A) Positive control; (B) Negative control; (C) CLET at concentration 1,000 µg/ml; (D) CLHE at concentration 500 µg/ml; (E) CLEA at concentration 200 µg/ml; (F) CLAQ at concentration 1,000 µg/ml. (40x magnification)
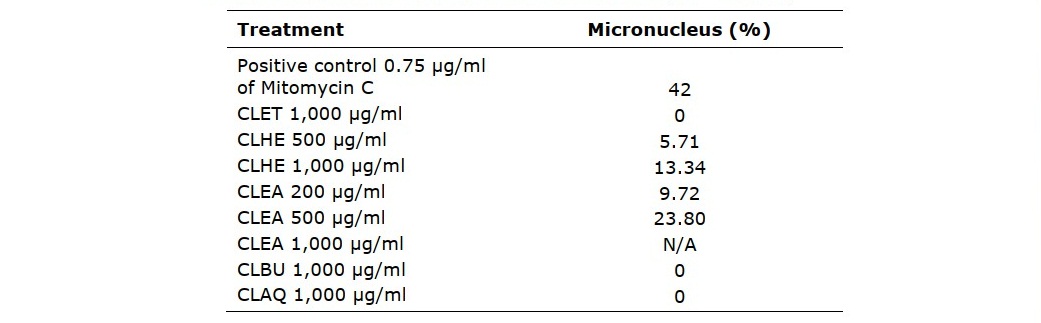
Genotoxicity of extracts from C. lentillifera by micronucleus test
Analyses of the effect on genetic material by the C. lentillifera extracts using the TK6 cell by micronucleus test (Figure 4) showed that in negative control the cell appeared to be binucleated (Figure 4B). In positive control the cell appeared to be binucleated with micronucleus at 42% (Figure 4A). Cells treated with CLET, CLBU, and CLAQ showed no micronucleus at 1,000 µg/ml, indicating that these three fractions were not toxic to the TK6 cells. On the other hand, CLHE and CLEA showed cells with micronucleus. For CLHE micronucleus was observed at concentrations 500 and 1,000 µg/ml with % of cells with micronucleus at 5.71 and 13.34%, respectively. For CLEA micronucleus was observed at concentrations 200 and 500 µg/ml with % of cells with micronucleus at 9.72 and 23.80%, respectively. These results confirmed that fractions CLET, CLBU, and CLAQ has no genotoxicity and did not cause any chromosomal damage, while CLHE and CLEA were safe at concentrations lower than 500 and 200 µg/ml, respectively.
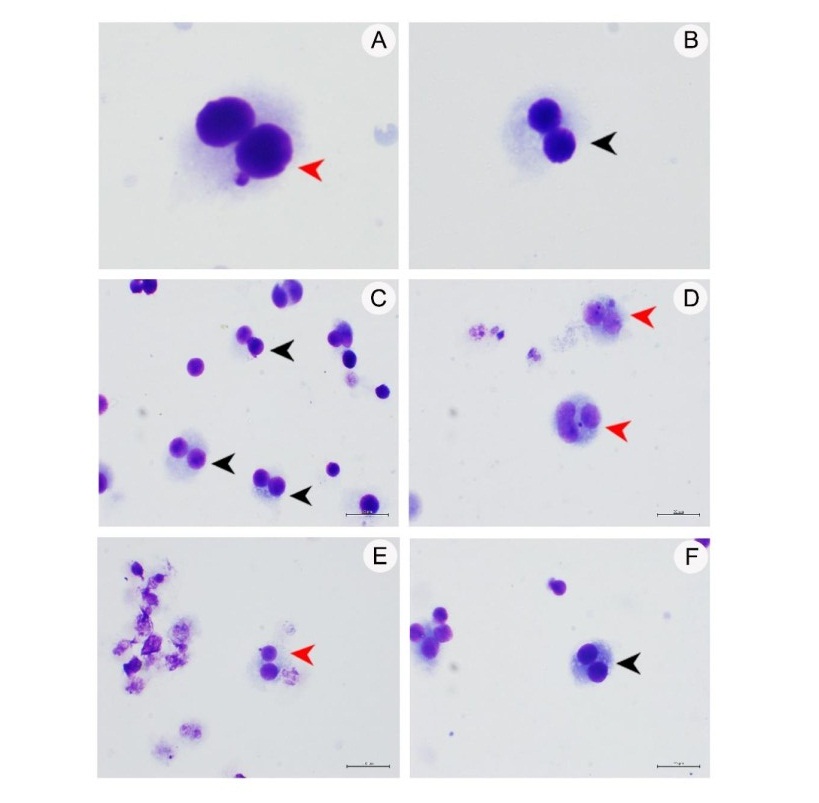
Figure 4. Genotoxicity as measured by micronucleus test indicated chromosome damage. (A) Positive control; (B) Negative control; (C) CLET at concentration 1,000 µg/ml; (D) CLHE at concentration 1,000 µg/ml; (E) CLEA at concentration 500 µg/ml; (F) CLAQ at concentration 1,000 µg/ml. Red and black arrow heads indicate binucleated cells with micronucleus and binucleated cells, respectively. (40x magnification)
Table 2. Genotoxicity measured by micronucleus assay showing % micronucleus in TK6 cells treated with C. lentillifera extracts for 24 h. (* P < 0.001)
Antioxidant activity of the extracts by DPPH and ABTS assays
In these assays, we have shown free radicals’ scavenging or antioxidant activity of C. lentillifera extracts by calculating and comparing EC50 values. The antioxidant activity of CLET fraction was between 1.21 ± 1.99 to 31.68 ± 1.65%, and CLHE was from 13.97 ± 6.68 to 38.27 ± 0.59%. The CLEA fraction showed highest antioxidant activity between 25.64 ± 1.84 to 91.25 ± 2.29%. The fractions showed the lowest antioxidant activity was CLBU between 4.50 ± 3.17 to 11.00 ± 5.70%. The CLAQ has moderate antioxidant activity at 3.04 ± 8.56% (Figure 5).
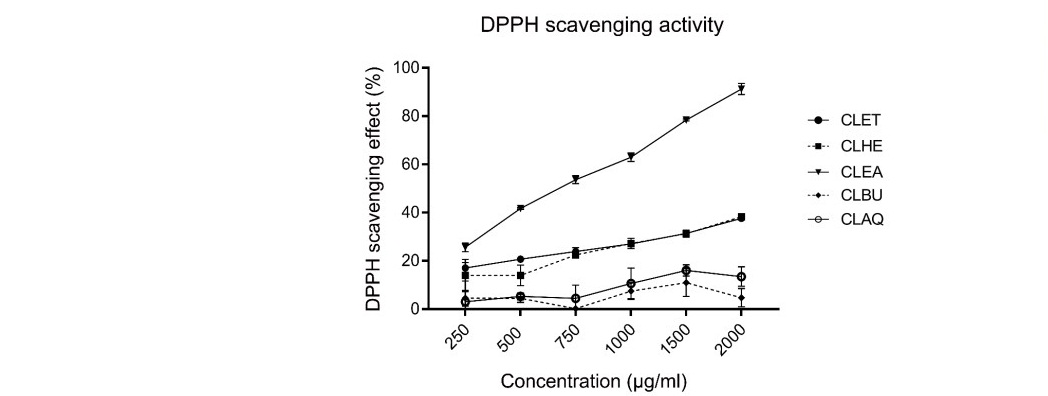
Figure 5. T DPPH scavenging effect of five fractions from C. lentillifera extracts. Each value was presented as mean ± standard error (n=3). The result was used to generate trendlines and for calculating the EC50 values.
The high percentage of inhibition of DPPH radicals was CLEA, CLHE, and CLET fractions, while CLAQ and CLBU fractions showed low antioxidant activity. The EC50 value of CLEA fraction was highest at 535 µg/ml. The EC50 values of CLHE and CLET fractions were 2,947.41 and 3,088.77 µg/ml. In contrast, the EC50 values of CLAQ and CLBU fractions were more than 5,000 µg/ml. Positive control (Vitamin C and quercetin) showed EC50 as 6.66 µg/ml and 2.37 µg/ml, respectively.
In ABTS assay, the five fractions of C. lentillifera extracts inhibit free radicals from ABTS radicals. The antioxidant activity of CLET was between 0.08 ± 2.74 to 46.46 ± 2.63%. The percentage of scavenging activity of CLET was increased when it increased concentration. The antioxidant activity of CLHE fractions was between 9.70 ± 1.81 to 40.41 ± 3.02%. The CLEA fraction showed the highest antioxidant activity between 28.75 ± 2.55 to 84.37 ± 0.97%. CLBU and CLAQ fractions showed low antioxidant activities at 21.99 ± 8.19 to 22.17 ± 1.56% and 18.26 ± 8.1 to 29.30 ± 4.15%, respectively (Figure 6).
The EC50 values of C. lentillifera extracts were calculated the equation of a line from %ABTS scavenging activity at 50% of the radical scavenging effect: CLEA, CLHE and CLET consecutively showed highest to lowest percentage of free radical scavenging activities with EC50 values at 712.42, 2,546.60 and 2,022.75 µg/ml, respectively. Positive control (Vitamin C and quercetin) showed EC50 as 4.82 µg/ml and 8.43 µg/ml, respectively.
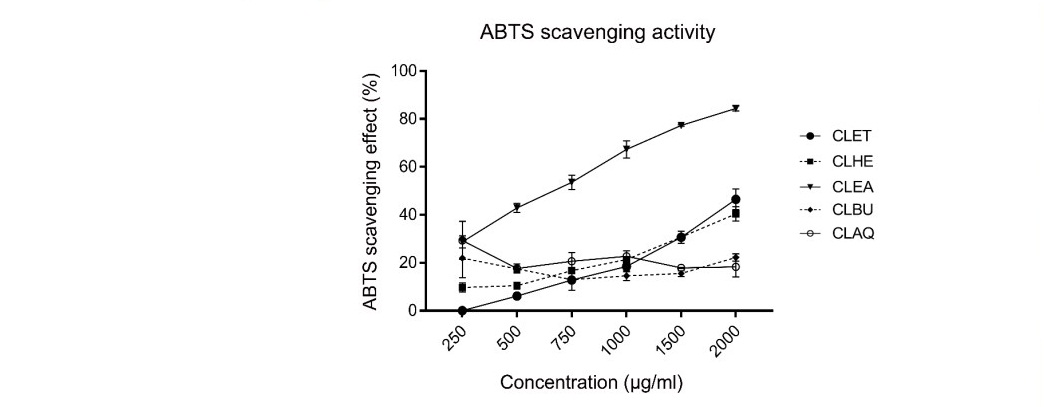
Figure 6. ABTS scavenging activities of five fractions from C. lentillifera extracts. Each value represents mean ± standard error (n=3). The data were used to generate trendlines and calculate the EC50 value.
DISCUSSION
We analyzed the cytotoxic potential of C. lentillifera extracts from low (10 μg/ml) to high (1,000 μg/ml) concentrations using standard MTT and LDH assays to confirm cytotoxicity in four cell lines. The results showed that RAW 264.7 and FL83B cells exhibited the lowest cell viability as these cells still underwent cell proliferation. Our results from MTT assay showed that fractions with lowest to the highest toxicity were CLAQ, CLET, CLBU, CLHE, and CLEA, respectively. Previous studies also reported that the CLET of the same seaweed did not showed cytotoxicity, including studies conducted in Japan at the highest concentration at 250 μg/ml (Sharma et al., 2015), in China at the highest concentration at 800 μg/ml (Sun et al., 2018) and in the Philippines at the highest concentration at 25 μg/ml (Sharma and Rhyu, 2014), and we have used an even higher concentration at 1,000 μg/ml. We found that CLHE and CLEA contains similar substances, including alkaloids, flavonoids, terpenoids, steroids, and glycoside, with tannin being present only in CLEA (our unpublished data). It has been reported that alkaloids, flavonoids, steroids, tannins, saponins, and terpenoids have toxicity around IC50 of 9.81 μg/ml (Singh et al., 2019). By contrast, we found the IC50 value of CLHE and CLEA to be higher at approximately 200 μg/ml which could be due to the lower concentrations of these substances in the extracts. Noticeably, CLHE and CLEA showed more toxicity than other fractions. Eventhough the above-mentioned substances were also present in CLET this fraction did not showed any cytotoxicity, probably because it contained these various substances in relatively low concentrations. Our results from LDH assay were more sensitive than MTT assay with IC50 of CLHE and CLEA being 544- 1025 μg/ml and 245-629 μg/ml, depending on cell types while CLET and CLAQ were nontoxic. This is because the MTT assay detected viable cells only (Fotakis and Timbrell, 2006), whereas LDH assay detected cells with damaged membranes which occurred earlier even before cell death. Nevertheless, the results from the two assays followed the same trend.
Comet assay has been used to assess genotoxicity as the result of DNA breakage. TK6 is a well-accepted cell for evaluating DNA damage in comet assay due to its high accuracy (90%) (Bajpayee et al., 2019; Bajpayee et al., 2006). Parameter employed in measuring DNA damage in cells included the percentage of DNA in the tail (% tail DNA), tail length, and tail moment calculated from fluorescence intensity observed under fluorescent microscope. Our data from comet assay showed that CLET, CLBU, and CLAQ at the highest concentration at 1,000 μg/ml showed no toxicity, while but CLHE and CLEA showed some genotoxicity at concentrations higher than 500 μg/ml and 200 μg/ml, respectively.
The micronucleus (MN) test has been used for assessing chromosomal damage in cells by micronucleus formation after cell division after treating TK6 cells for 24 h. The percentage of binucleated cells with micronucleus indicated that fractions or chemicals have cytostatic action on mitogenic activity. This assay was used to confirm comet assay. It was found that CLET, CLBU, and CLAQ did not damage chromosomes but CLHE and CLEA caused significant damage at concentrations higher than 500 μg/ml and 200 μg/ml, respectively.
The antioxidant activity of C. lentillifera extracts were measured by DPPH and ABTS assays. The ethanol, butanol, and water are polar solvents whose dielectric constants are 24.3, 18, and 78.5, respectively. They were used to extract hydrophilic antioxidants such as phenolics, flavonoids, and anthocyanins (Xu et al., 2017). On the other hand, hexane is non-polar solvent with dielectric constant at 1.9. It was used for extracting lipo-soluble antioxidants such as carotene, lycopene, and xanthine. The ethyl acetate is a polar aprotic solvent with intermediate dielectric constant and polarity (He et al., 2016; Xu et al., 2017). It is used for extracting both water-soluble as well as lipid-soluble antioxidants. Furthermore, the ethanol is commonly used for extraction because it has medium polarity, that can both dissolve water- and lipid-soluble antioxidants (Xu et al., 2017) and usually employed at the first stage of extraction.
The data from this study showed CLEA fraction showed the highest antioxidant activity followed by CLHE, CLET, CLAQ, and CLBU. CLEA contained alkaloids, tannins, terpenoids, and glycosides. As well, the phytochemicals in CLHE (alkaloids, flavonoids, terpenoids, and glycosides) were similar to CLEA fraction, thus antioxidant activity of CLHE and CLEA fractions were very close. In contrast, The CLET was extracted with ethanol, that contained chlorophyll, alkaloids, flavonoids, phenols, tannins, terpenoids, steroids, glycosides and saponins, at low concentrations in consequence of low antioxidant activity. Finally, the CLBU and CLAQ fractions extracted with high polar, the phytochemicals of these include phenols. The CLBU and CLAQ fractions showed the lowest antioxidant effect because they may contain low concentration of phenol.
In previous studies, C. lentillifera was often extracted by using ethanol, chloroform, and water. The high antioxidant activities are expressed by fractions in non- polar solvent. The chloroform extract showed significantly higher antioxidant activity when compared to methanol and water extracts. The EC50 of DPPH scavenging effect with chloroform, methanol, and water extract were about 2.20 ± 0.10, 9.74 ± 0.59, and 81.55 ± 4.22 mg/ml (Yap et al.,2019). Compared to our result, The DPPH radical scavenging activity obtained from chloroform extract was lower than CLEA but higher than CLHE and CLET. Thus, the extraction methods affected involve the antioxidant activity. C. lentillifera was extracted with ethanol by thermal-drying and freeze-drying methods. The freeze-drying method showed a higher antioxidant effect than thermal- drying methods (Nguyen et al., 2011).
CONCLUSION
In summary, we have investigated cytotoxic, genotoxicity, and antioxidant activity of different extracts from C. lentillifera (CLET, CLHE, CLEA, CLBU, CLAQ) in in vitro models of different cell types including hepatocyte, fibroblast, macrophage and keratinocyte in order to assess the nontoxic concentrations of the extracts from this seaweed that could safely be utilized in the food, cosmetic and drug development industries.
ACKNOWLEDGEMENT
The authors thank the Faculty of Allied Health Sciences, Burapha University for providing instruments.
CONFLICT OF INTEREST
The authors declare that they hold no competing interests.
REFERENCES
AbouZid, S.F., Ahmed, O.M., Ahmed, R.R., Mahmoud, A., Abdella, E., and Ashour, M.B. 2014. Antihyperglycemic effect of crude extracts of some Egyptian plants and algae. J Med Food. 17: 400-406.
Apostolova, N. and Victor, V.M. 2015. Molecular strategies for targeting antioxidants to mitochondria: therapeutic implications. Antioxid Redox Signal. 22: 686-729.
Arenajo, A.R., Ybanez, A.P., Ababan, M.M.P., Villajuan, C.E., Lasam, M.R.M., Young, C.P., and Reyes, J.L.A. 2017. The potential anticoagulant property of Caulerpa lentillifera crude extract. Int J Health Sci (Qassim). 11: 29-32
Arulselvan, P., Fard, M.T., Tan, W.S., Gothai, S., Fakurazi, S., Norhaizan, M.E., and Kumar, S.S. 2016. Role of Antioxidants and Natural Products in Inflammation. Oxid Med Cell Longev. 2016: 5276130.
Bajpayee, M., Kumar, A., and Dhawan, A. 2019. The comet assay: assessment of in vitro and in vivo DNA damage. Methods Mol Biol. 2031: 237-257.
Bajpayee, M., Pandey, A.K., Zaidi, S., Musarrat, J., Parmar, D., Mathur, N., and Dhawan, A. 2006. DNA damage and mutagenicity induced by endosulfan and its metabolites. Environ Mol Mutagen. 47: 682-692.
Collins, A.R., Oscoz, A.A., Brunborg, G., Gaivao, I., Giovannelli, L., Kruszewski, M., and Štětina, R. 2008. The comet assay: topical issues. Mutagenesis. 23: 143-151.
FAO. 2020. The State of World Fisheries and Aquaculture 2020. Sustainability in action. Rome.
Fotakis, G. and Timbrell, J.A. 2006. In vitro cytotoxicity assays: comparison of LDH, neutral red, MTT and protein assay in hepatoma cell lines following exposure to cadmium chloride. Toxicol Lett. 160: 171-177.
He, T., Pachfule, P., Wu, H., Xu, Q., and Chen, P. 2016. Hydrogen carriers. Nature Reviews Materials. 1: 16059.
Maeda, R., Ida, T., Ihara, H., and Sakamoto, T. 2012. Induction of apoptosis in MCF-7 cells by β-1, 3-xylooligosaccharides prepared from Caulerpa lentillifera. Biosci Biotechnol Biochem. 76: 1032-1034.
Matanjun, P., Mohamed, S., Mustapha, N.M., Muhammad, K., and Ming, C.H. 2008. Antioxidant activities and phenolics content of eight species of seaweeds from north Borneo. Journal of Applied Phycology. 20: 367.
Nguyen, V.T., Ueng, J.P., and Tsai, G.J. 2011. Proximate composition, total phenolic content, and antioxidant activity of seagrape (Caulerpa lentillifera). J Food Sci. 76: C950-C958.
Palanisamy, S., Rajasekar, P., Vijayaprasath, G., Ravi, G., Manikandan, R., and Prabhu, N.M. 2017. A green route to synthesis silver nanoparticles using Sargassum polycystum and its antioxidant and cytotoxic effects: an in vitro analysis. Materials Letters. 189: 196-200.
Sharma, B.R. and Rhyu, D.Y. 2014. Anti-diabetic effects of Caulerpa lentillifera: stimulation of insulin secretion in pancreatic β-cells and enhancement of glucose uptake in adipocytes. Asian Pac J Trop Biomed. 4: 575-580.
Sharma, B.R., Kim, H.J., and Rhyu, D.Y. 2015. Caulerpa lentillifera extract ameliorates insulin resistance and regulates glucose metabolism in C57BL/KsJ-db/db mice via PI3K/AKT signaling pathway in myocytes. J Transl Med. 13: 62.
Sharma, B.R., Kim, H.J., and Rhyu, D.Y. 2017. Caulerpa lentillifera inhibits protein- tyrosine phosphatase 1B and protects pancreatic beta cell via its insulin mimetic effect. Food Sci Biotechnol. 26: 495-499.
Singh, R.K., Ranjan, A., Srivastava, A.K., Singh, M., Shukla, A.K., Atri, N., and Singh, S.K. 2019. Cytotoxic and apoptotic inducing activity of Amoora rohituka leaf extracts in human breast cancer cells. J Ayurveda Integr Med. 0975-9476.
Sun, Y., Gong, G., Guo, Y., Wang, Z., Song, S., Zhu, B., and Jiang, J. 2018. Purification, structural features and immunostimulatory activity of novel polysaccharides from Caulerpa lentillifera. Int J Biol Macromol. 108: 314-323.
Walum, E., Clemedson, C., and Ekwall, B. 1994. Principles for the validation of in vitro toxicology test methods. Toxicology in vitro. 8: 807-812.
Yap, W.F., Tay, V., Tan, S.H., Yow, Y.Y., and Chew, J. 2019. Decoding Antioxidant and Antibacterial Potentials of Malaysian Green Seaweeds: Caulerpa racemosa and Caulerpa lentillifera. Antibiotics (Basel, Switzerland), 8(3), 152.
Xu, D.P., Li, Y., Meng, X., Zhou, T., Zhou, Y., Zheng, J., and Li, H.B. 2017. Natural Antioxidants in Foods and Medicinal Plants: Extraction, Assessment and
OPEN access freely available online
Chiang Mai University Journal of Natural Sciences [ISSN 16851994]
Chiang Mai University, Thailand https://cmuj.cmu.ac.th
Supawadee Osotprasit1, Tepparit Samrit1, Athit Chaiwichien1, Narin Changklungmoa1, Krai Meemon2, Nakorn Niamnont3, Preeyanuch Manohong3, Kunwadee Noonong4, Montakan Tamtin5, Prasert Sobhon2, and Pornanan Kueakhai1,*
1 Faculty of Allied Health Sciences, Burapha University, Chonburi 20131, Thailand.
2 Department of Anatomy, Faculty of Science, Mahidol University, Bangkok 10400, Thailand.
3 Department of Chemistry, Faculty of Science, King Mongkut's University of Technology Thonburi, Bangkok 10140, Thailand.
4 School of Allied Health Sciences, Walailak University, Nakhonsithammarat 80160, Thailand.
5 Coastal Aquatic Feed Research Institute, Coastal Fisheries Research and Development Bureau, Department of Fisheries, Phetchaburi 76000, Thailand.
Corresponding author: Pornanan Kueakhai, E-mail: earn_patho@hotmail.com; pornanan@go.buu.ac.th
Total Article Views
Editor: Korakot Nganvongpanit, Chiang Mai University, Thailand
Article history:
Received: August 31, 2020;
Revised: March 15, 2021;
Accepted: March 18, 2021;
Published online: April 5, 2021

