
In Silico Targeting DENV2's Prefusion Envelope Protein by Several Natural Products' Bioactive Compounds
Arief Hidayatullah1, Wira Eka Putra*, Sustiprijatno, Galuh Wening Permatasari, Wa Ode Salma, Diana Widiastuti, Hendra Susanto, Bayyinatul Muchtaromah, Dewi Ratih Tirto Sari, Febby Nurdiya Ningsih, Muhammad Fikri Heikal, Alyana Mahdavikia Rosyada Yusuf, and Aliyya Suci ArizonaPublished Date : 2021-04-01
DOI : https://doi.org/10.12982/CMUJNS.2021.059
Journal Issues : Number 3, July-September 2021
Abstract Dengue caused by the dengue virus (DENV) is a severe health problem in tropical regions such as Southeast Asia, especially Indonesia. Indonesian have used rhizome as traditional medicine for 1300 years. This study investigated the compounds from Kaempferia galanga, Curcuma longa, Zingiber officinale, Curcuma aeruginosa, Curcuma zanthorrhiza, Alpinia galanga, and Allium sativum as antivirals agents, explicitly targeting the DENV envelope protein to inhibit viral fusion. This study involved 121 bioactive compounds and DENV2's prefusion envelope protein. The virtual screening and molecular docking were done through occupied the Lipinski rule of five checker (http://www.scfbio-iitd.res.in/software/drugdesign/lipinski.jsp) and AutoDock Vina (https://pyrx.sourceforge.io/) respectively. The top nine compounds with the strongest binding affinity were galangin, kampferide, demetoxy curcumin, bisdemethoxycurcumin, β-selinene, 6-(hydroxymethyl)-1,4,4-trimethylbicyclo[3.1.0]hexan-2-ol, piperine, estra-1,3, 5(10)-trien-17β-ol, and curcumin. These compounds' affinity values were significantly lower, around 45-62%, than chloroquine. Most of them interact with the kl hairpin and hydrophobic pocket formed by residues Val130, Leu135, Phe193, Leu198, and Phe279of critical domains that can interfere with the conformational change and rearrangement of protein dimer in the post-fusion stage. This study suggested that the galangin, demethoxycurcumin, and bisdemethoxycurcumin are considered the most potential compounds to be developed as anti-prefusion E DENV2 low-affinity and intense interaction with those.
Keywords: DENV2, envelope protein, in silico, viral fusion, viral infection
Funding: This study was funded by PNBP Universitas Negeri Malang (Wira Eka Putra).
Citation: Hidayatullah, A., Putra, W.E., Sustiprijatno, Permatasari, G.W., Salma, W.O., Widiastuti, D., Susanto, H., Muchtaromah, B., Sari, D.R.T., Ningsih, F.N., Heikal, M.F., Yusuf, A.M.R., and Arizona, A.S. 2021. In Silico Targeting DENV2's Prefusion Envelope Protein by Several Natural Products' Bioactive Compounds. CMU J. Nat. Sci. 20(3): e2021059.
INTRODUCTION
Dengue virus, also known as DENV, is a positive-sense single-stranded RNA virus from the genus Flavivirus with four known serotypes (DENV1-4). DENV causes dengue fever, one of the most dangerous neglected tropical diseases and mosquito-borne diseases in the world (Alen & Schols, 2012; Shah et al., 2013; Warkentien, 2016; World Health Organization & Department of Control of Neglected Tropical Diseases, 2017; Tahir et al., 2018). DENV virus's protein consists of two main groups: structural protein groups and nonstructural proteins. Structural proteins are responsible for the structure of mature DENV virions, while nonstructural proteins are responsible for the replication process of DENV in target cells. The structural components of DENV consist of capsid (C), membrane (M), and envelope (E), while the nonstructural components consist of seven nonstructural proteins (NS1, 2a, 2b, 3, 4a, 4b, and 5) (Norazharuddin & Lai, 2018). Among the DENV structural proteins, an envelope protein (E) is studied as a target protein in antiviral development. The DENV2 E protein is a class II fusion protein that works through receptor-mediated, clathrin-dependent endocytosis triggered by a low pH environment (Butrapet et al., 2011; Smit et al., 2011; Alen & Schols, 2012; de Wispelaere et al., 2018). In the early infection stage, E protein of DENV changes the fusion unit conformation and viral membrane protein fuses with the target (Alen & Schols, 2012; Klein et al., 2013; Shah et al., 2013; Harrison, 2015; Rouvinski et al., 2017). This process is crucial for infection mechanism of DENV, and might be used for antiviral targeting. Nowadays, some studies explored compounds that potential as antivirals target. Arbidol as antivirals compound prevented H1N1 and H3N2 infections, and Enfuvirtide inhibited the HIV fusion to CD4+ cells via CCR5 receptors (Lange et al., 2013; Tian et al., 2018; Vankadari, 2020; Wang et al., 2020).
Indonesia has at least around 9,000 species of plants that predicted to have medicinal properties. However, only a ninth of them have been widely used, mostly as herbal ingredients. Only about 500 species have been used as phytopharmaca materials (Salim & Munadi, 2017; Jadid et al., 2020). The Indonesian people are familiar with using certain types of plants as traditional medicines, carried out from generation to generation, often through oral lore. This oral tradition from generation to generation has resulted in the lack of written records regarding these plant groups' initial use in Indonesian culture. Several records show that the Indonesian people have carried out plants as traditional medicine before the 8th century AD, depicted on reliefs at Borobudur Temple and several other ancient manuscripts (Subositi & Wahyono, 2019).
In Indonesian culture, plants' use as traditional medicines is considered adequate and has minimal side effects, although there is still no adequate scientific explanation regarding these traditional medicines (Jennifer & Saptutyningsih, 2015; Sumayyah & Salsabila, 2017). Some communities assumed that the traditional medicinal herb is more effective than drugs and continually generated to their family and other communities (Hakim, 2015; Parwata, 2016; Badan Pengawas Obat dan Makanan, 2019; Fakultas Kedokteran, Kesehatan Masyarakat dan Keperawatan Universitas Gajah Mada, 2020). Research conducted by the Indonesian Ministry of Health shows that 95.6% of 59.12% communities assumed the traditional herbal medicine is beneficial for health (Siswanto, 2012; Andriati & Wahjudi, 2016). The most widely used medicinal plants in Indonesian culture are the rhizome group, such as turmeric, galangal, temulawak, and ginger (Beers, 2012; Woerdenbag & Kayser, 2014; Hakim, 2015; Kristianto et al., 2020). Some research suggests that several bioactive compounds from these group can potentially develop as a potent antiviral for certain viruses such as Human papillomavirus (HPV), influenza, human respiratory syncytial virus (HRSV), and even dengue itself (Dao et al., 2012; Abdel-Moneim et al., 2013; Chang et al., 2013; Ichsyani et al., 2017).
There are several studies had been conducted regarding potential bioactive compounds that may act as antiviral against DENV (Loaiza-Cano et al., 2021). The nonstructural protein, especially the NS2B/NS3 protease and NS5, are the most commonly used target proteins in the antiviral development process because they have well-mapped enzymatic activity for viral polyprotein and RNA synthesis (Norazharuddin & Shin, 2018; Jennings & Parks, 2020). These proteins are also well-conserved among DENV serotypes (Moghadamtousi et al., 2015; García et al., 2017; Tahir et al., 2018; Silva et al., 2019). A conserved protein is a protein whose amino acid sequence has fewer amino acid replacements or is more likely to substitute amino acids with similar biochemical properties among all DENV serotypes represented by a percentage (Kairys & Fernandes, 2007; Sousounis et al., 2012).
In previous in silico research, halishigamide A was predicted to be the strongest candidate against DENV2 NS5 due to low binding affinity value (Hidayatullah et al., 2020). The development of the DENV antiviral is starting to consider other target proteins besides nonstructural proteins, such as DENV’s envelope protein, which is playing the critical role during the early infection stage. Hence, this study performed to screening on several types of rhizomes that commonly used in Indonesia, such as Kaempferia galanga, Curcuma longa, Zingiber officinale, Curcuma aeruginosa, Curcuma zanthorrhiza, Alpinia galanga, and Allium sativum to find the bioactive compounds that could be developed as promising antivirals by explicitly targeting the DENV envelope protein to inhibit viral fusion. This study expected to be fundamental of future study of anti-DENV discovery, especially for natural bioactive compound.
MATERIALS AND METHODS
Ligands and protein data mining
The protein that is projected to be the target in this study is the envelope protein (E) of the DENV serotype 2, the most critical protein in the DENV fusion mechanism. Specifically, the E protein conformation used is the prefusion conformation that occurs at the earliest stage of the DENV fusion process, so the disruption that occurs due to interactions with potential compounds occurs in the earliest phase of the viral fusion process (Tian et al., 2018). The 3D protein E structure used was obtained from the primary database, the RSCB PDB (https://www.rcsb.org/) (1OKE) with the PDB file format that compatible with AutoDock Vina (Y. Modis et al., 2003). The 3D structure is a homodimer structure with a sequence length of 394 residues with 2-acetamido-2-deoxy-beta-D-glucopyranose as the native ligand on Asn67 and Asn153 residues (Alen & Schols, 2012). The target protein is optimized by removing the native ligand from the amino acid sequence using PyMol.
The compounds data are obtained through a data mining process based on active compounds found in several medicinal plants' rhizomes commonly used in Indonesia. There are seven rhizomes used as a source of potential compounds in this study. They are Kaempferia galanga, Curcuma longa, Zingiber officinale, Curcuma aeruginosa, Curcuma zanthorrhiza, Alpinia galanga, and Allium sativum (Karyawati, 2011; Dao et al., 2012; Abdel-Moneim et al., 2013; Chang et al., 2013; Yan et al., 2014; Theanphong et al., 2015; Ichsyani et al., 2017; Munda et al., 2018; Sugita et al., 2018; Mao et al., 2019; Shang et al., 2019). We determined the 121 bioactive compounds from these selected sources will be mined at PubChem (https://pubchem.ncbi.nlm.nih.gov/). The drug compound used as a control in this study was chloroquine (CID: 2719), which, based on several studies, showed the potential for DENV fusion inhibition (Mao, 2019; Sugita, 2018). All data mining compounds are 3D structural data in the SDF format compatible with AutoDock Vina and can be converted into AutoDock ligand format.
Ligands virtual screening
A data mining process precedes this research method. The data mining process is divided into two parts: the protein data mining process and the potential compound data mining process. All compounds obtained from the data mining process were tested using the Lipinski Rule of Five parameters. This test was carried out by the Supercomputing Facility for Bioinformatics and Computational Biology, IT Delhi (http://www.scfbio-iitd.res.in/software/drugdesign/lipinski.jsp) server (Jayaram et al., 2012). Compounds that passed the test must meet all the Lipinski parameters' criteria without any tolerance. These compounds can be declared to have pharmacological properties chemically and physically suitable for drugs, especially for humans' oral consumption. Compounds that pass the Lipinski test are then optimized using the PyRx program, which is integrated with the OpenBabel GUI by minimizing the energy of these compounds with the aim that the protein-ligand interaction distance results are accurate to the level of quantum chemical calculations so that they are biologically close to the actual state (Mirzaei et al., 2015).
Molecular docking process
The docking process is performed using AutoDock Vina (https://pyrx.sourceforge.io/) (Trott & Olson, 2009). All components must be converted into AutoDock Macromolecule and AutoDock Ligand (pdbqt) format. After the target protein macromolecules and compounds are selected, the docking coordinates are determined. Because this research is exploratory, the docking coverage is the entire structure of the E protein. The molecular coverage area (in Angstrom) is X: 39.8415, Y: 56.3282, and Z: 149.8127, and the center at X: -12.7517, Y: 69.0073, Z: 24.4419. The docking process is repeated three times to avoid abnormal data. The main docking results are the affinity in kcal/mol, the binding site location, and the protein-ligand interaction, known through the visualization process.
Visualization of Docking Analysis
The process of visualizing the results is carried out in two stages. The first stage is the 3D visualization of target protein complexes and potential natural compounds. This visualization process uses PyMOL (https://pymol.org/2/). This primary purpose of visualization is to provide a general visual description of potential natural compounds' binding sites on the target protein. The second stage of visualization is 2D visualization to determine the interaction between target proteins and potential natural compounds in 2D (Wallace et al., 1995). This visualization stage uses LigPlot+ version 2.1. (https://www.ebi.ac.uk/thornton-srv/software/LigPlus/). This 2D visualization process aims to determine the interaction between the target protein and the potential compound and the interaction distance between the two components.
Protein target of each compound and Network Analysis
To further explore the protein target of each compound, we performed the prediction target analysis. The protein target were then analyse its interaction using protein-protein interaction database called STRING. The Gene Ontology (GO), including cellular compounds (CC), molecular function (MF), and biological process (BP) was generated from STRING algorithm (https://string-db.org/).
RESULTS
Virtual screening of 121 compounds showed that nine compounds have potential activity as drugs. The galangin, kampferide, demetoxy curcumin, bisdemethoxycurcumin, β-selinene, 6-(hydroxymethyl)-1,4,4-trimethylbicyclo[3.1.0] hexan-2-ol, piperine, estra-1,3,5(10)-trien-17β-ol, and curcumin interacted with E protein of DENV with the strong binding affinity. These natural compounds' affinity value is range -7.7 to -8.6 kcal/mol, 45-62% lower than the control (Chloroquine; -5.3 kcal/mol) (Figure 1).
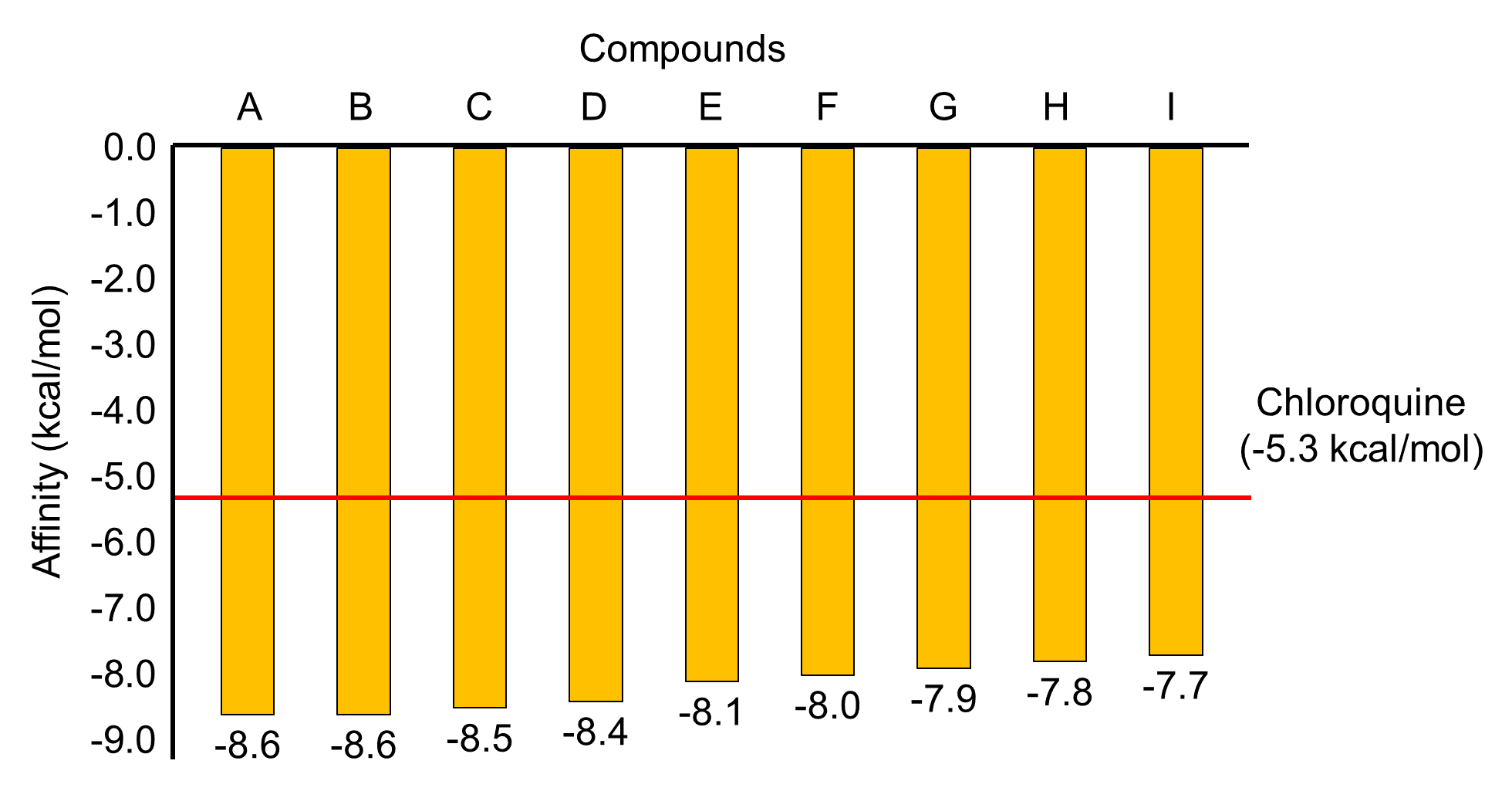
Figure 1. The comparison of the affinity value of top nine compounds with chloroquine based on the docking results of the prefusion DENV2 E protein. (A) galangin, (B) kampferide, (C) demetoxy curcumin, (D) bisdemethoxycurcumin, (E) β-selinene, (F) 6-(hydroxymethyl)-1,4,4-trimethylbicyclo[3.1.0]hexan-2-ol, (G) piperine, (H) curcumin, (I) Estra-1,3,5(10)-trien-17β-ol.
The 2D visualization performed that top nine compounds against prefusion DENV2 E protein by hydrophobic interactions (Table 1). Demetoxy curcumin, 6-(hydroxymethyl)-1,4,4-trimethylbicyclo[3.1.0]hexan-2-ol, estra-1,3,5(10)-trien-17β-ol, and chloroquine interact with residues on both monomers; whereas kaempferide, bisdemethoxycurcumin, β-selinene, piperine, and curcumin each only interacted with the residue on a monomer, either monomer A or B. 2D visualization showed that the most conserved residues of the two monomers were Ile270, Leu198, and Leu207. The results also indicated that 50% of the target protein residues interacted with more than one potential natural compounds.
The 3D and 2D visualization show that the top nine compounds and control are concentrated on the three binding pockets on the E protein structure, namely A, B-B,' C, and D (Figure 2, 3 and 4). The A site is occupied by demetoxy curcumin, and estra-1,3,5(10)-trien-17β-ol, the B site is occupied by curcumin and β-selinene, while the counterpart occupied by galangin, kaempferide, bisdemethoxycurcumin, and piperine; the C site is occupied by 6-(hydroxymethyl)-1,4,4-trimethylbicyclo[3.1.0]hexan-2-ol; and the D site occupied by chloroquine. Thus, the A and C sites formed between monomer A and B, while the B-B' site formed either on monomer A or B. The use of an apostrophe (') in the binding site notation is due to its formation in the same residue complex but in the opposite monomer. All potential compounds interact with at least one residue in the kl hairpin complex, except chloroquine. Chloroquine interacts at binding pocket D, isolated from the other compound, but barely similar with the rest of the compounds in the A site with three similar residues. They are Lys247, Asp154, and Arg2.
Table 1. Docking and 2D visualization result of top nine compounds against prefusion DENV2 E protein.
|
No. |
Compound |
Source |
DG Average (kcal/mol) |
Amino Acids Residue |
Interaction (Å) |
|
1. |
Galangin (CID: 5281616) |
Alpinia galanga |
-8.6 |
Ile270(B); Phe193(B); Leu207(B); Val130(B); Thr48(B); Leu135(B); Glu49(B); Gln200(B); Leu198(B); Ala205(B); Gln271(B); Ala50(B) |
Hydrophobic interaction |
|
|
|
|
|
Gln271(B) |
Hydrogen bond (2.90) |
|
|
|
|
|
Ala50(B) |
Hydrogen bond (2.88) |
|
2. |
Kampferide (CID: 5281666) |
Alpinia galanga |
-8.6 |
Ile270(B); Leu198(B); Leu207(B); Phe193(B); Thr280(B); Thr48(B); Glu49(B); Leu135(B); Gln200(B); Ala205(B); Gln271(B); Ala50(B) |
Hydrophobic interaction |
|
|
|
|
|
Gln271(B) |
Hydrogen bond (2.88) |
|
|
|
|
|
Ala50(B) |
Hydrogen bond (2.83) |
|
3. |
Demethoxy curcumin (CID: 5324476) |
Curcuma longa |
-8.5 |
Ser273(A); Leu278(A); Lys241(B); Pro243(B); His244(B); Ile46(A); Arg2(A); Glu44(A); Leu45(A); Asn276(A); Gln248(B); Asp249(B); Lys247(B); Leu277(A); Asp154(A); His27(A); Gly28(A). |
Hydrophobic interaction |
|
|
|
|
|
Lys247(B) |
Hydrogen bond (2.93) |
|
|
|
|
|
Leu277(A) |
Hydrogen bond (2.71) |
|
|
|
|
|
Asp154(A) |
Hydrogen bond (2.82) |
|
|
|
|
|
His27(A) |
Hydrogen bond (3.18) |
|
|
|
|
|
Gly28(A) |
Hydrogen bond (3.09) |
|
4. |
Bisdemethoxycurcumin (CID: 5315472) |
Curcuma longa |
-8.4 |
Leu191(B); Gly281(B); Val130(B); Leu207(B); Ile270(B); Thr280(B); Ala205(B); Gln271(B); Leu135(B); Thr268(B); Leu198(B); Phe279(B); Gly190(B); Gln200(B) |
Hydrophobic interaction |
|
|
|
|
|
Gly190(B) |
Hydrogen bond (3.22) |
|
|
|
|
|
Gln200(B) |
Hydrogen bond (3.25) |
|
5. |
β-Selinene (CID: 442393) |
Curcuma aeruginosa |
-8.1 |
Leu135(A); Ile270(A); Thr48(A); Ala50(A); Val130(A); Leu198(A); Leu207(A); Thr280(A) |
Hydrophobic interaction |
|
6. |
6-(Hydroxymethyl)-1,4,4-trimethylbicyclo[3.1.0]hexan-2-ol (CID: 538067) |
Zingiber officinale |
-8.0 |
Thr262(B); Leu253(A); Lys204(B); His261(B); Met272(B); Thr239(A); Trp206(B); Leu264(B); Glu269(B); Thr265(B) |
Hydrophobic interaction |
|
7. |
Piperine (CID: 638024) |
Zingiber officinale |
-7.9 |
Ile270(B); Thr280(B); Leu207(B); Phe193(B); Leu198(B); Val130(B); Thr48(B); Leu277(B); Gln271(B); Ser274(B) |
Hydrophobic interaction |
|
|
|
|
|
Gln200(B) |
Hydrogen bond (3.10) |
|
8. |
Curcumin (CID: 969516) |
Curcuma longa |
-7.8 |
Thr268(A); Leu207(A); Gly190(A); Phe279(A); Leu135(A); Leu277(A); Gln271(A); Gln200(A); Leu198(A); Thr48(A); Ile270(A); Thr280(A); Phe193(A); Thr189(A); Gly281(A); Leu191(A) |
Hydrophobic interaction |
|
9. |
Estra-1,3,5(10)-trien-17β-ol (CID: 228944) |
Zingiber officinale |
-7.7 |
Glu44(A); Leu277(A); Leu278(A); Lys247(B); Asn276(A); Pro243(B); Lys246(B); His244(B); Ile46(A) |
Hydrophobic interaction |
|
|
|
|
|
Ile46(A) |
Hydrogen bond (2.75) |
|
10. |
Chloroquine (CID: 2719) |
Antiviral drug (control) |
-5.3 |
Val97(A); Ile113(A); Arg99(A); Ser72(A); Thr70(A); Lys247(A); Asp154(B); Asn103(A); Asp98(A); Arg2(B); Ile6(B) |
Hydrophobic interaction |
Galangin, the potential compound with the best affinity, is predicted to interact with two crucial residue complexes. They are kl hairpin (Ile270, Gln271) and hydrophobic pocket (Phe193, Val130, Leu135). The interaction with Ile270(B) is a hydrophobic with the main core structure. In contrast, the interaction with Gln271(B) consists of two interactions, hydrophobic interaction and hydrogen bonding with moderate strength with the hydroxyl group on C8. Meanwhile, the hydrophobic pocket interaction entirely occurs through a stable hydrophobic contact with the central core structure and the hydroxyl group at C5. Kaempferide, which has the same affinity value as galangin and interacted with the critical residue of E protein. β-selinene bound to the kl hairpin residue Ile270(A) and the hydrophobic pocket residues Leu198(A), Val130(A), and Leu135(A) of DENV E protein. The same pattern also showed in piperine, which interacted by hydrophobic contact with kl hairpin residues Ile270(B), Gln271(B), Ser274(B), and Leu277(B). Piperin also interacted with hydrophobic pockets residue Phe193(B) and Leu198(B) of DENV E protein, occurs between the heterocyclic ring and the methylenedioxyphenyl ring in both terminals.
Demethoxy curcumin bound to three critical residues on the kl hairpin, the Ser273(A), Leu277(A), and Leu278(A), and one residue on the DI-DIII linkage, the His27(A). The interactions between the Ser273(A) and Leu278(A) residues are hydrophobic contacts, especially with the hydroxyl group on the C8 and the ether group on the C7. While the hydrophobic contact makes up the interaction with Leu277(A), it is also supported by the hydrogen bonding with moderate strength that interacts with the main structure and the hydroxyl group at the C1. The interaction with the DI-DIII linkage in His27(A) involves hydrophobic contact and moderate hydrogen bond with the hydroxyl group on the C12. Although demethoxy curcumin also interacts with B monomer, the critical residue is wholly bound to A monomer.
Bisdemethoxycurcumin is a potent compound with the fourth-highest affinity value, which is predicted to interact with three critical domains; the kl hairpin, hydrophobic pocket, and hinge. Interaction with kl hairpin (Ile270(B), Gln271(B), and Phe279(B)), hydrophobic pocket (Val130(B) and Leu198(B)), and hinge (Leu191(B)) of E DENV protein. Curcumin interacted with kl hairpin of E DENV protein (Leu277(A)), and hydrophobic pockets (Leu135(A), Phe193(A),Val130(B)).
6-(Hydroxymethyl)-1,4,4-trimethylbicyclo[3.1.0]hexan-2-ol is the compound with the sixth-highest affinity value, also the only compound that occupies the binding site C. This compound interacts with one critical, Met272(B), which is part of the kl hairpin through hydrophobic contact with the compound's main structure. However, when we observed the binding site's position, this compound is precisely sandwiched between two EDII domains, near the twofold axis.
The interaction pattern of the estra-1,3,5(10)-trien-17β-ol shows this compound only interacts with the critical residue of the kl hairpin, the Leu277(A), Leu278(A), and Asn276(A) through hydrophobic contact which occurs with the main structure of this compound. Like demethoxy curcumin, although this compound interacts with both monomers, the interaction with the critical residues only occurs with monomer A.
Chloroquine, as control, which had the lowest affinity value, showed different residue complexes' interaction compared to all potential compounds. This compound interacts with the fusion loop at Arg99(A), Asp98(A), and Asn103(A). The interactions are entirely hydrophobic contact between these residues and the main bicyclic structure, [5-(diethylamino)pentan-2-yl] amino group, and chlorine.

Figure 2. An overview of the binding sites based on the top nine compounds' 3D visualization results with prefusion DENV2 E protein. (A) galangin, (B) kampferide, (C) demetoxy curcumin, (D) bisdemethoxycurcumin, (E) β-selinene, (F) 6-(hydroxymethyl)-1,4,4-trimethylbicyclo[3.1.0]hexan-2-ol, (G) piperine, (H) curcumin, (I) Estra-1,3,5(10)-trien-17β-ol, and (J) chloroquine as control.
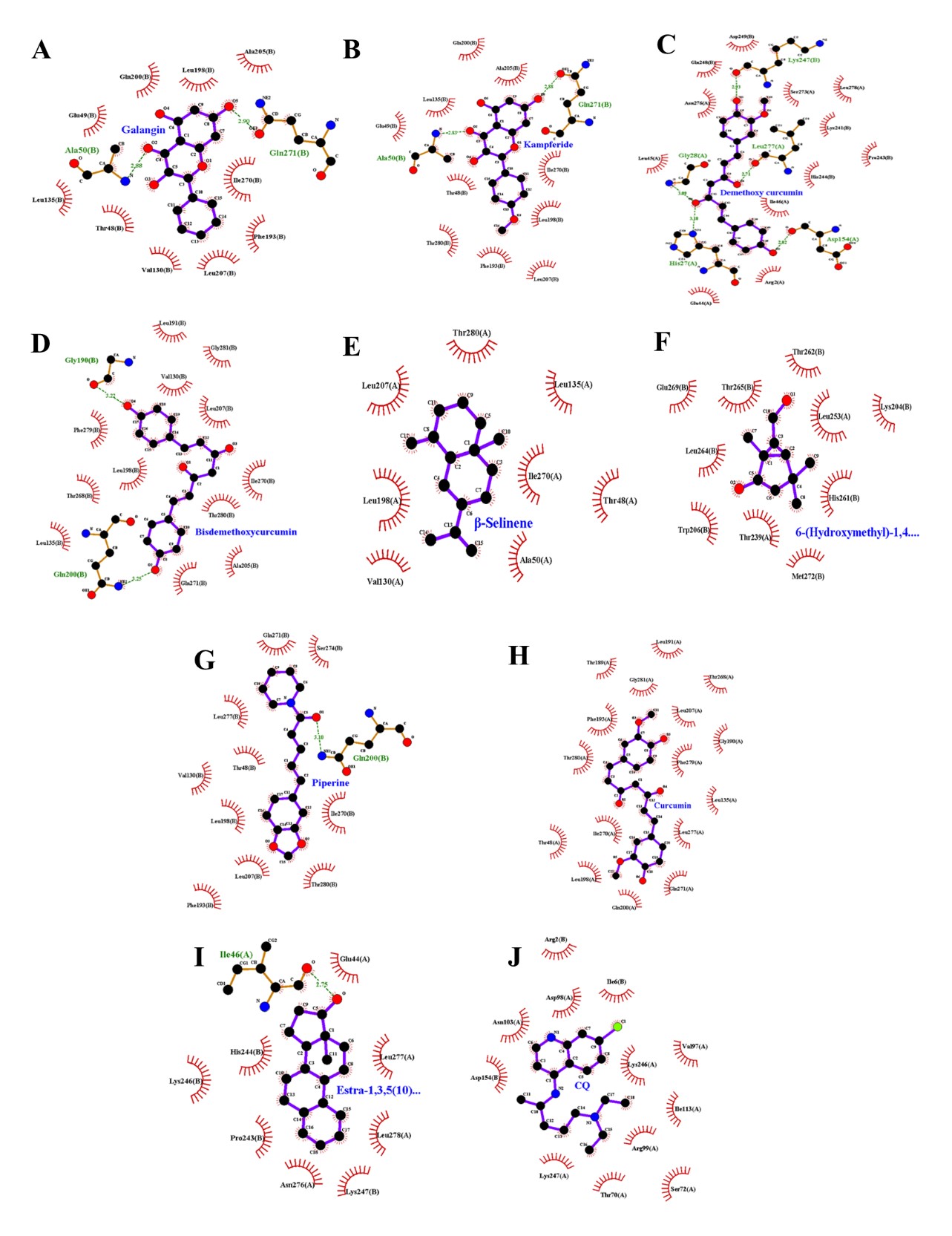
Figure 3. The interaction between the top nine compounds molecule with the prefusion DENV2 E protein based on the 2D visualization results. (A) galangin, (B) kampferide, (C) demetoxy curcumin, (D) bisdemethoxycurcumin, (E) β-selinene, (F) 6-(hydroxymethyl)-1,4,4-trimethylbicyclo[3.1.0]hexan-2-ol, (G) piperine, (H) curcumin, (I) Estra-1,3,5(10)-trien-17β-ol, and (J) chloroquine as control.
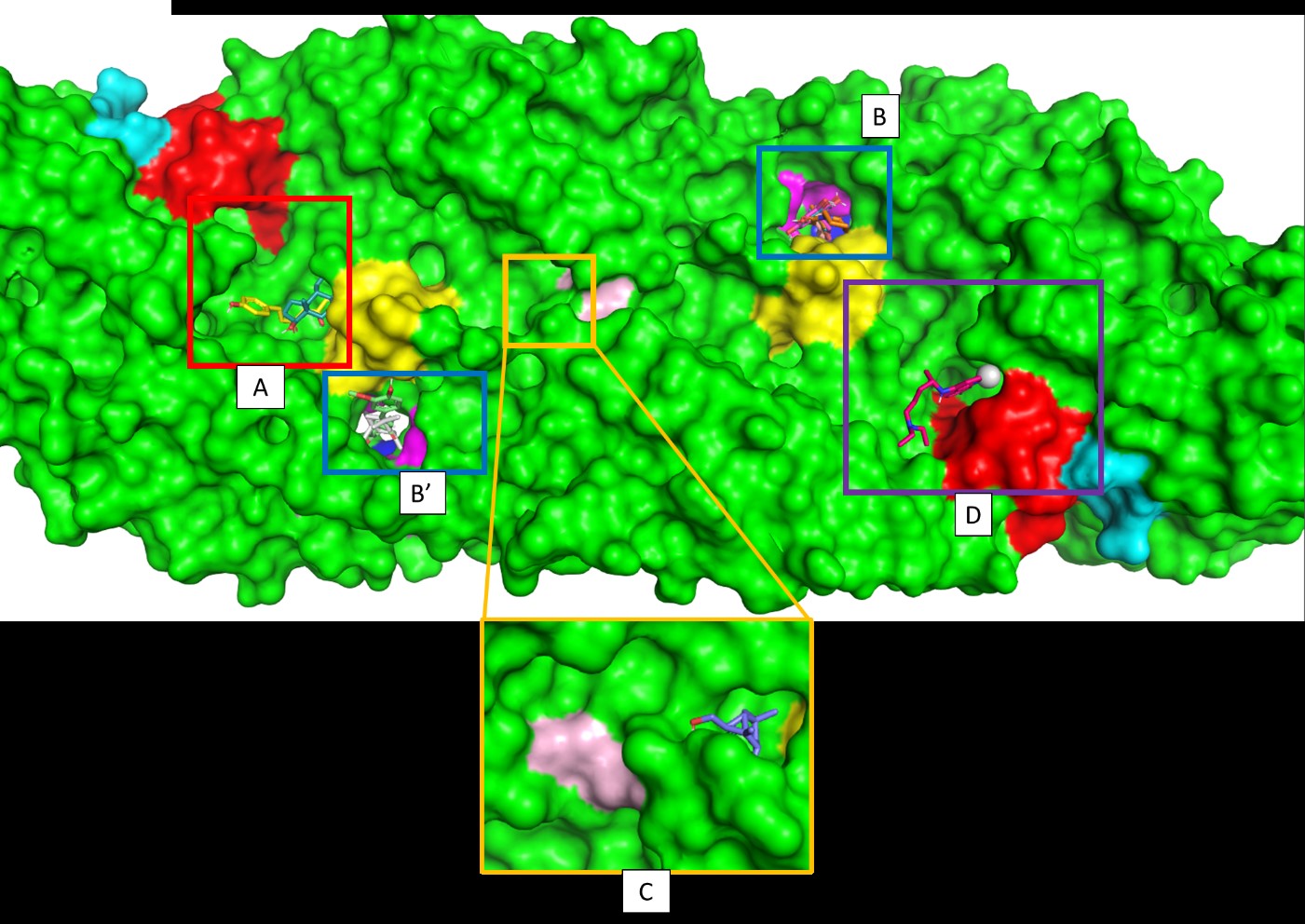
Figure 4. The top nine compounds and control occupy the approximate binding site. The A site is occupied by demethoxy curcumin and estra-1,3,5(10)-trien-17β-ol. The B site is occupied by curcumin and β-selinene. The B' site is occupied by galangin, kaempferide, bisdemethoxycurcumin, and piperine. The C site occupied by 6-(Hydroxymethyl)-1,4,4-trimethylbicyclo[3.1.0]hexan-2-ol. Finally, the D site occupied by chloroquine. The fusion loops showed in red, the protein hinge region showed in blue, the kl hairpin showed in yellow, the twofold axis showed in light magenta, the A strand showed in cyan, and the hydrophobic pocket showed in magenta. The AB loop cannot be showed in this picture
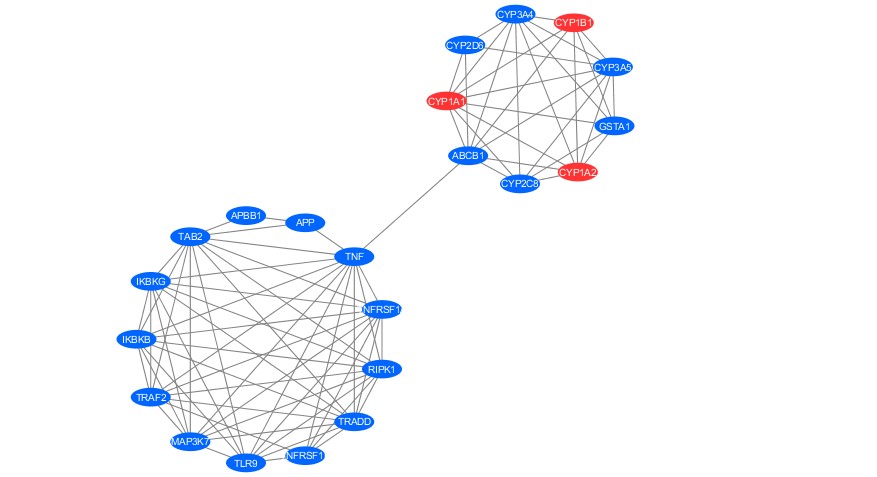
Figure 5. Networking analysis of active compound (Blue, circle) and protein target of Galangin and Kaempferide (Red, circle).
To deepen the hypothesis related to the effectivity of those active compounds for prefusion, protein target prediction and PPI analysis were performed. The protein target of Galangin and Kaempferide involving CYB1B1, CYP1A1, and CYP1A2 with 100% score of precision. Initially, we got 26 protein targets from 10 compounds (including control) showed in Table 1. However, to obtain the valid target, we filtered the finding based on its precision score, which is allowed only for 100% score. Twelve protein targets were retrieved, including APP, TLR9, TNF, CYP2D6, CYP3A5, GSTA1, ABCB1, CYP2C8, CYP3A4, CYP1B1, CYP1A1 and CYP1A2. Those targets were then predict its function and interaction with the nearest protein using networking analysis and gene ontology (GO).
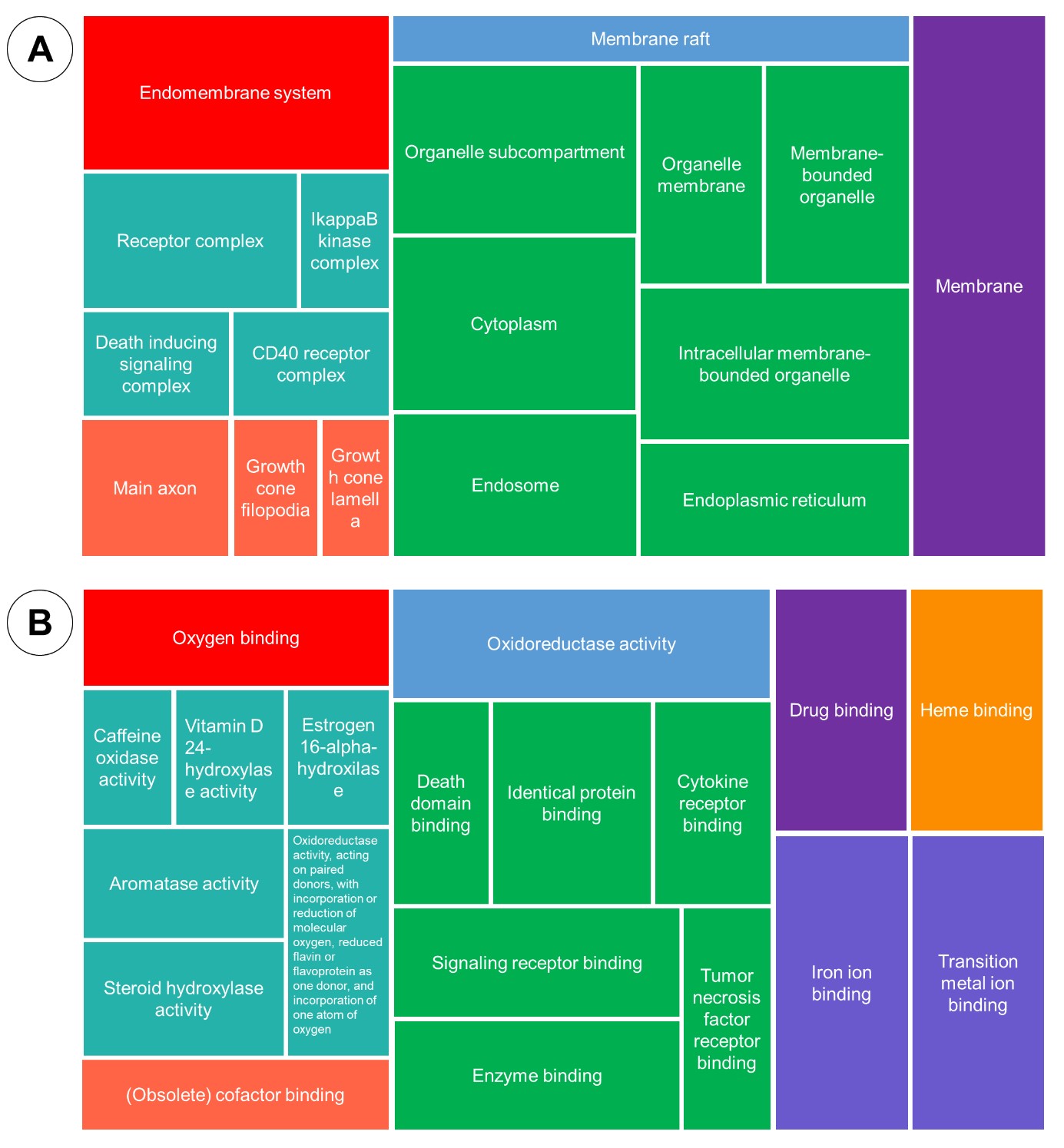
Figure 6. Gene Ontology analysis: cellular component (A), molecular function (B).
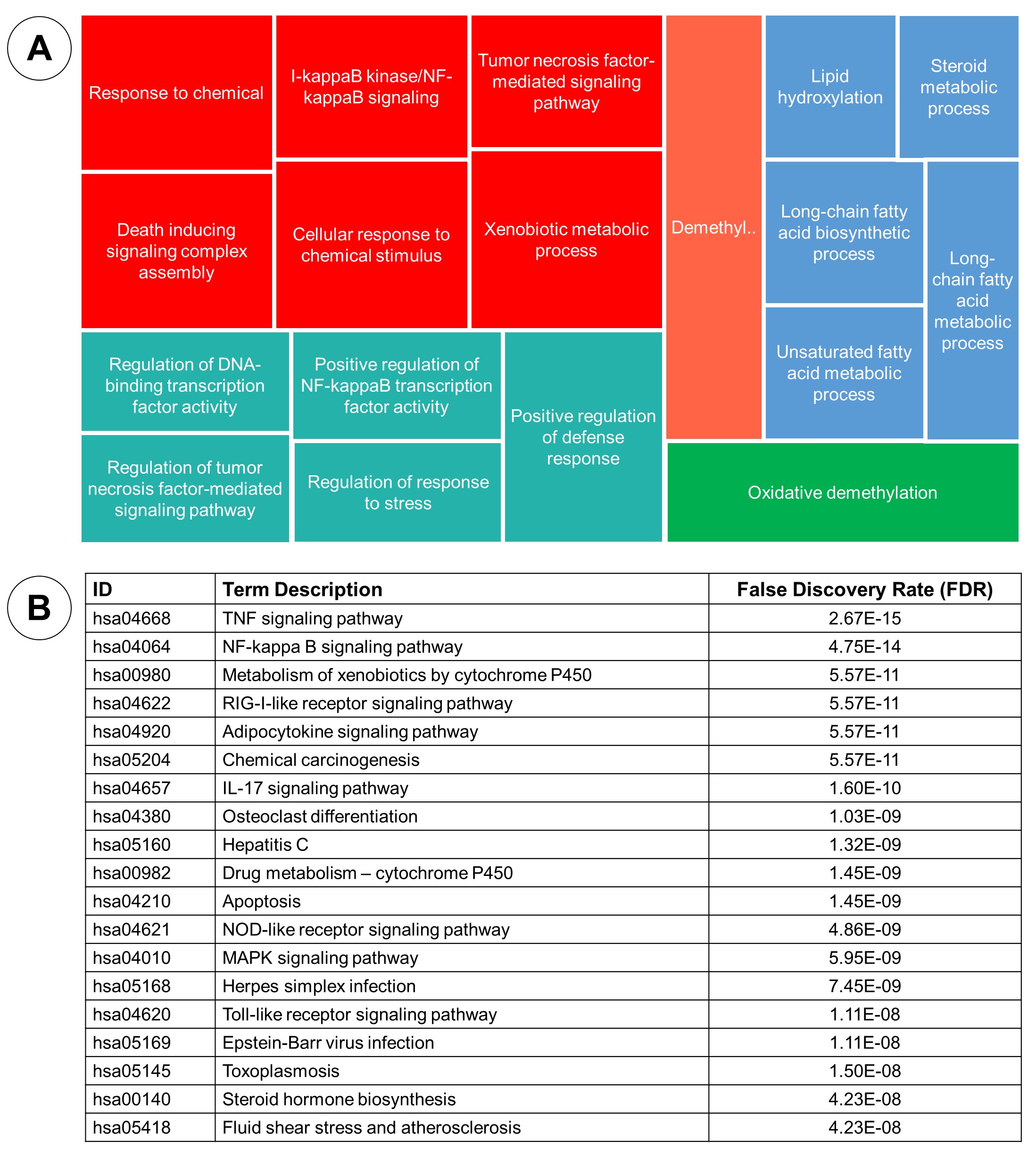
Figure 7. Gene Ontology analysis: biological process (A), and KEGG enrichment (B).
The PPI analysis formed two main clusters related to the protein target of active compounds. The target of Galangin and Kaempferide generated a cluster with others target protein, including CYP2D6, CYP3A4, CYP3A5, GSTA1, CYP2C8, and ABCB1. Those proteins hypothetically have function related to the perfusion of DENV virus through cytochrome associated pathway.
The enrichment of biological process pointed out terms related to the response to chemical stimulus, demethylation, several terms associated with metabolic process. While molecular function enrichment stated some terms related to the binding oxygen, iron, cofactor, enzyme, drug and protein. The cellular component showed location of the target, including membrane, endoplasmic reticulum and cytoplasm. Lastly, the KEGG enrichment revealing the term related to the TNF signaling, NF-Kappa B signaling, metabolism of xenobiotics through cytochrome P450 and any other terms rated to the virus infection.
DISCUSSION
The protein envelope located on the surface of the DENV is one of the crucial proteins that play a role in the initial mechanism of infection with Flavivirus such as DENV because it facilitates receptor recognition and fusion of the viral membrane with target cells such as dendritic cells (Degrève et al., 2012; Klein et al., 2013). Mature DENV 2 virions are dimer protein complexes that lay flat in the virus surface and must do some conformational changes to interact with the cell receptor and mediate the fusion process (Alen & Schols, 2012; Klein et al., 2013). This phase is known as the prefusion state. Mature DENV 2 new virions will undergo a conformational change to a post-fusion state triggered by certain conditions, especially in a low pH environment (Alen & Schols, 2012; Klein et al., 2013; Shah et al., 2013). We choose prefusion state conformation because it is mainly on this conformation until the infection's early stage. These proteins have several important domains that play a role in infection mechanisms and can be used as target domains disrupted by antiviral compounds. Some of these critical protein complexes include fusion loops (98-109), protein hinge region (191), kl hairpin (270-279), twofold axis (255,259), AB loop (314-319), A strand (310-313), and a hydrophobic pocket (130, 135, 193, 198, and 279) (Klein et al., 2013; Rouvinski et al., 2017; Anasir et al., 2020).
The critical protein complexes with the most significant chance of being disrupted by the potential compounds tested in this study are kl hairpin and hydrophobic pocket. The dominant interaction between these compounds and kl hairpin is hydrophobic contact. Hydrophobic interactions performed stronger than other intermolecular interactions involved hydrogen bonds or Van der Waals bonds, indicating that the ligand-protein complexes were relatively stable (Jeffrey, 1997; Atkins & Paula, 2010).
kl β-hairpin has known for its act in initiating the conformational changes at low pH to form fusion component trimers (Modis et al., 2003). Any binding of small molecules on the ligand-binding pocket in opened kl hairpin conformation may prevent fusion in low pH (Mir et al., 2016).
However, demethoxy curcumin has one hydrogen bond with Leu277(A) with a donor-acceptor distance around 2.71Å, which indicates that the bond has a moderate intensity and mostly electrostatic (Jeffrey, 1997). An additional hydrogen bond occurs between demethoxy curcumin and Leu277(A), which is also thought to stabilize the ligand-protein complex. Theoretically, the ligand-protein complex formed is more stable than piperine, and curcumin only interacts through hydrophobic contact.
On the other hand, demethoxy curcumin also has the lower affinity than piperine and curcumin. Theoretically, demethoxy curcumin has a higher tendency for spontaneous interactions and stable interaction between these compounds (Murcko & Ajay, 1995; Trott & Olson, 2009; Pantsar & Poso, 2018). The interactions that occurred between demethoxy curcumin and the kl hairpin in envelope domain II (EDII), is predicted to interfere with the conformational change and rearrangement of protein dimer into the post-fusion stage, which is indispensable for the process of virus entry to target cells through the membrane fusion mechanism (Yorgo Modis et al., 2005; Alen & Schols, 2012; Anasir et al., 2020). Therefore, demethoxy curcumin is probably one of the most potent compounds that can inhibit the kl hairpin complex.
The next critical protein complex most likely to be disrupted by the most potent compounds is the hydrophobic pocket formed by residues 130, 135, 193, 198, and 279. Among the top nine compounds that interact with this protein complex are galangin, kaempferide, bisdemethoxycurcumin, β-selinene, piperine, and curcumin; galangin is a compound that has the most interactions with residues in the hydrophobic pocket, which interact with four residues. Thus, this compound interacts with 80% of the residue that makes up the hydrophobic pocket. This high interaction ratio is supported by a stable hydrophobic contact that makes up the ligand-protein complex also supported by the affinity value of galangin. The affinity value of galangin is 62% lower than the control, which indicates its interaction and hydrophobic pocket has a higher tendency for spontaneous interactions. Thus, the complex formed is more stable than others, including control (Murcko & Ajay, 1995; Trott & Olson, 2009; Pantsar & Poso, 2018). This makes galangin may disrupt the hydrophobic pocket efficiently. Blocking or disrupting the hydrophobic pocket by antiviral compounds will interfere with the conformation change process of E protein during the fusion process (Modis et al., 2003) The hydrophobic pocket plays key role during conformation changes since it serves as a hinge in the protein activation mechanism that stimulated by a low pH environment so that it can affect the pH threshold also indirectly affect the virulence of the virus (Alen & Schols, 2012; Anasir et al., 2020).
In addition to those dominant complexes, two potential compounds are known to interact with Leu191, a hinge region that connects EDI-EDII and facilitates the 30° rotation required by EDII when doing a conformational change from prefusion into the post-fusion state. The hinge region's influence is also closely related to the kl hairpin previously described (Yorgo Modis et al., 2004). These potential compounds are curcumin and bisdemethoxycurcumin that interact with Leu191 on the monomers A and B, respectively, through hydrophobic interactions. Both compounds also interact with kl hairpin, so the conformational change inhibition mechanism will likely occur via those two regions. Hence, these two compounds' inhibition potential is also promising considering that they have relatively stable interaction through hydrophobic contact and have a lower affinity than control.
According to the binding site, 6-(hydroxymethyl)-1,4,4-trimethylbicyclo [3.1.0]hexan-2-ol is the only compound that binds near the twofold axis region of E protein, although the results show that these compounds interact with one kl hairpin residue. The interaction near the twofold axis of the E protein dimer can interfere with the disulfide bond between the two monomers through its axis so that it is projected to disrupt the conformation of the E protein (Rouvinski et al., 2017). However, this compound does not interact exactly on the twofold axis residue, so this assumption still needs further investigation.
There is one crucial region that does not interact with potential compounds but interacts with chloroquine as control. These regions are the fusion loops. This fusion loop is buried at the dimer interface in the mature virion but exposed during fusion. Several sources state that the fusion loop is the most critical region in the initial mechanism of dengue infection because it functions to link the virus to the target cell membrane and serves as an anchor in the protein conformation changing process, which causes the bend of the virus and cell membranes to a certain distance so the membrane fusion process can occur (Klein et al., 2013; Anasir et al., 2020). Chloroquine was predicted\ inhibited viral fusion mechanism by disrupting the fusion loop and prevented the virus attaching to the target cell membrane. However, because chloroquine has the lowest affinity than the top nine compounds, chloroquine complexes with fusion loops are the most unstable ligand-protein complexes and the lowest chance of spontaneous formation theoretically (Murcko & Ajay, 1995; Trott & Olson, 2009; Pantsar & Poso, 2018).
Galangin, kampferide, demethoxy curcumin, bisdemethoxycurcumin and curcumin are belong to the flavonoid group that has been known for its activity as anti-bacterial and anti-viral. The chemical structure of Galangin and kampferide shows the existence of dual fused aromatic rings (Ring A and C), that reported in previous research to posses antiviral activity against NS2B-NS3 complex of the dengue virus (Sarwar et al., 2018; Lalani & Poh, 2020). In addition, galangin and kampferide that show the best affinity to E protein of DENV 2, consist of more hydroxyl group number compared to other flavonoid compound in this study. Though more hydroxyl groups number give lower hydrophobicity that limit flavonoids transport through biological membranes, sometimes certain hydroxyl group-rich-flavonoids give higher activity in in vitro study. It may happened due to the balance of hydrophobicity and electronic delocalization on the strength of hydroxylation assignment (Wang et al., 2018; Zou et al., 2020). Thus, the fused aromatic rings that conserved in most flavonoid also hydroxylation in their skeleton structure may responsible to antiviral activity on E protein. However, the relationship of chemical structure from this five compounds with their antiviral action specifically on DENV 2 E protein need to be investigated more.
We then performing the GO enrichment analysis to further reveal the Galangin and Kaempferide active compound capacity in more big picture. The KEGG enrichment showing terms related to the TNF, NF Kappa B signaling pathway and metabolism via Cytochrome pathway. It might be related to the potential of Galangin and Kaempferide to inhibit the DENV infection process via oxidative stress. The reactive oxygen species (ROS) is begin to accumulate once the DENV infected the patient, so that it elevate the level of proinflammatory cytokines. The location of ROS secretion is in the plasma membrane of NADPH as the results of infection as a defence mechanism/antiviral mechanism. Next, the IRF3-STAT1 and NF Kappa B were induced and regulate the inflammatory cytokine expression. This action was then continued with NOX-derived ROS which trigger a p53 protein, so that the propagation of virus limited (Olaigner et al., 2014). Thus, we discovered that active compounds of Galangin and Kaempferide paying crucial role as an antioxidant to stop the ROS production and DENV infection.
CONCLUSION
Based on the tests and analyses conducted, we conclude that the compounds considered the most potential to be developed as anti-E DENV2 in the prefusion state are galangin, demethoxy curcumin, and bisdemethoxycurcumin. Galangin was chosen because it has the best affinity and has strong interaction with a hydrophobic pocket. Demethoxy curcumin is also chosen because it has an affinity that is only 0.2 kcal/mol higher than galangin and has the best interaction with the kl hairpin. Lastly, demethoxycurcumin was also chosen because of its low-affinity value and interaction with the kl hairpin and simultaneously with the hinge region. Further studies are needed to confirm this study's prediction and further develop these compounds as antiviral compounds that are explicitly targetting the DENV E protein.
ACKNOWLEDGMENTS
This study was funded by PNBP Universitas Negeri Malang (Wira Eka Putra). All authors thanks Universitas Negeri Malang for support this study.
AUTHOR CONTRIBUTIONS
Arief Hidayatullah and Wira Eka Putra conceptualised the central research idea, provided the theoretical framework, wrote and revised the article. Arief Hidayatullah, Muhammad Fikri Heikal, Alyana Mahdavikia Rosyada Yusuf, and Aliyya Suci Arizona carried out the experiments and discussion. Sustiprijatno, Galuh Wening Permatasari, Wa Ode Salma, Diana Widiastuti, Hendra Susanto, Bayyinatul Muchtaromah, Dewi Ratih Tirto Sari, Febby Nurdiya Ningsih provided the discussion, theoretical framework, and revised the article. All authors anchored the review, revisions and approved the article submission.
CONFLICT OF INTEREST
The authors declare that they hold no competing interests.
REFERENCES
Abdel-Moneim, A., Morsy, B.M., Mahmoud, A.M., Abo-Seif, M.A., and Zanaty, M.I. 2013. Beneficial therapeutic effects of Nigella sativa and/or Zingiber officinale in HCV patients in Egypt. EXCLI Journal. 12: 943–955.
Alen, M.M.F. and Schols, D. 2012. Dengue Virus Entry as Target for Antiviral Therapy. Journal of Tropical Medicine. 1–13.
Anasir, M.I., Ramanathan, B., and Poh, C.L. 2020. Structure-Based Design of Antivirals against Envelope Glycoprotein of Dengue Virus. Viruses. 12: 367.
Andriati, A. and Wahjudi, R.M.T. 2016. Tingkat penerimaan penggunaan jamu sebagai alternatif penggunaan obat modern pada masyarakat ekonomi rendah-menengah dan atas. Masyarakat, Kebudayaan dan Politik. 29: 133.
Atkins, P. and Paula, J. de. 2010. Physical Chemistry for the Life Sciences (Second Edition). Oxford University Press.
Badan Pengawas Obat dan Makanan. 2019. Peraturan Badan Pengawas Obat dan Makanan Nomor 32 tahun 2019 Tentang Persyaratan Keamanan dan Mutu Obat Tradisional. Badan Pengawas Obat dan Makanan.
Beers, S.-J. 2012. Jamu: The ancient Indonesian art of herbal healing. Tuttle Publishing.
Butrapet, S., Childers, T., Moss, K.J., Erb, S.M., Luy, B.E., Calvert, A.E., Blair, C.D., Roehrig, J.T., and Huang, C.Y.-H. 2011. Amino acid changes within the E protein hinge region that affect dengue virus type 2 infectivity and fusion. Virology, 413: 118–127.
Chang, J.S., Wang, K.C., Yeh, C.F., Shieh, D.E., and Chiang, L.C. 2013. Fresh ginger (Zingiber officinale) has antiviral activity against human respiratory syncytial virus in human respiratory tract cell lines. Journal of Ethnopharmacology. 145: 146–151.
Dao, T.T., Nguyen, P.H., Won, H.K., Kim, E.H., Park, J., Won, B.Y., and Oh, W.K. 2012. Curcuminoids from Curcuma longa and their inhibitory activities on influenza A neuraminidases. Food Chemistry. 134: 21–28.
de Wispelaere, M., Lian, W., Potisopon, S., Li, P.-C., Jang, J., Ficarro, S.B., Clark, M.J., Zhu, X., Kaplan, J.B., Pitts, J.D., et al. 2018. Inhibition of flaviviruses by targeting a conserved pocket on the viral envelope protein. Cell Chemical Biology. 25: 1006-1016.e8.
Degrève, L., Fuzo, C.A., and Caliri, A. 2012. Extensive structural change of the envelope protein of dengue virus induced by a tuned ionic strength: Conformational and energetic analyses. Journal of Computer-Aided Molecular Design. 26: 1311–1325.
Fakultas Kedokteran, Kesehatan Masyarakat dan Keperawatan Universitas Gajah Mada. (2020, June 5). Obat tradisional di era pandemi covid-19 [University Web]. https://fk.ugm.ac.id/obat-tradisional-di-era-pandemi-covid-19/
Farias, K.J.S., Machado, P.R.L., Junior, R.F. de A., Aquino, A.A. de, and Fonseca, B.A.L. da. 2014. Chloroquine interferes with dengue-2 virus replication in U937 cells. Microbiology and Immunology. 58: 318–326.
García, L.L., Padilla, L., and Castaño, J.C. 2017. Inhibitors compounds of the flavivirus replication process. Virology Journal. 14.
Hakim, L. 2015. Rempah dan Herba Kebun Pekarangan Rumah Masyarakat: Keragaman, Sumber Fitofarmaka dan Wisata Kesehatan-Kebugaran. Diandra Creative.
Harrison, S.C. 2015. Viral membrane fusion. Virology, (2015), 498–507.
Hidayatullah, A., Putra, W.E., Salma, W.O., Muchtaromah, B., Permatasari, G.W., Susanto, H., Widiastuti, D., and Kismurtono, M. 2021. Discovery of drug candidate from various natural products as potential novel dengue virus nonstructural protein 5 (NS5) inhibitor. Chiang Mai University Journal of Natural Sciences. 20:1-17
Ichsyani, M., Ridhanya, A., Risanti, M., Desti, H., Ceria, R., Putri, D.H., Sudiro, T.M., and Dewi, B.E. 2017. Antiviral effects of Curcuma longa L. against dengue virus in vitro and in vivo. IOP Conference Series: Earth and Environmental Science, 101: 012005.
Jadid, N., Kurniawan, E., Himayani, C.E.S., Andriyani, Prasetyowati, I., Purwani, K.I., Muslihatin, W., Hidayati, D., & Tjahjaningrum, I.T.D. 2020. An ethnobotanical study of medicinal plants used by the Tengger tribe in Ngadisari village, Indonesia. PLoS ONE. 15.
Jayaram, B., Singh, T., Mukherjee, G., Mathur, A., Shekhar, S., and Shekhar, V. 2012. Sanjeevini: A freely accessible web-server for target directed lead molecule discovery. BMC Bioinformatics. 13: S7.
Jeffrey, G.A. 1997. An introduction to hydrogen bonding. Oxford University Press.
Jennifer, H., and Saptutyningsih, E. 2015. Preferensi individu terhadap pengobatan tradisional di indonesia. Jurnal Ekonomi dan Studi Pembangunan. 16: 16.
Jennings, M.R., and Parks, R.J. 2020. Curcumin as an antiviral agent. Viruses. 12: 1242.
Kairys, V. and Fernandes, M.X. 2007. SitCon: Binding site residue conservation visualization and protein sequence-to-function tool. International Journal of Quantum Chemistry, 107: 2100–2110.
Karyawati, A.T. 2011. Aktivitas antivirus simian retrovirus serotype-2 (SRV-2) dari ekstrak meniran (Phyllanthus niruri) dan temu lawak (Curcuma Xanthorrhiza). Jurnal Penelitian Sains, 14, Article 3.
Klein, D. E., Choi, J. L., and Harrison, S.C. 2013. Structure of a dengue virus envelope protein late-stage fusion intermediate. Journal of Virology, 87: 2287–2293.
Kristianto, S., Batoro, J., Widyarti, S., and Sumitro, S. (2020, October 31). Exploration and economic value of medicinal plants as traditional herbal ingredients in bangselok, madura, indonesia.
Lalani, S. and Poh, C.L. 2020. Flavonoids as antiviral agents for enterovirus A71 (EV-A71). Viruses, 12: 184.
Lange, D.C., Secemsky, E.A., Ho, J.E., and Hsue, P.Y. 2013. Chapter 48—Manifestations, mechanisms, and treatment of HIV-Associated cardiovascular disease. In E. M. Antman & M. S. Sabatine (Eds.), Cardiovascular Therapeutics: A Companion to Braunwald's Heart Disease (Fourth Edition) (pp. 728–737). W.B. Saunders.
Loaiza-Cano, V., Monsalve-Escudero, L.M., Filho, C.d.S.M.B., Martinez-Gutierrez, M., and Sousa, D.P.d. 2021. Antiviral role of phenolic compounds against dengue virus: a review. Biomolecules, 11: 11.
Mao, Q.-Q., Xu, X.-Y., Cao, S.-Y., Gan, R.-Y., Corke, H., Beta, T., and Li, H.-B. 2019. Bioactive compounds and bioactivities of ginger (Zingiber officinale Roscoe). Foods, 8:185.
Mir, A., Ismatullah, H., Rauf, S., Niazi, U.H.K. 2016. Identification of bioflavonoid as fusion inhibitor of dengue virus using molecular docking approach. Informatics in Medicine Unlocked, 3: 1-6.
Mirzaei, H., Zarbafian, S., Villar, E., Mottarella, S., Beglov, D., Vajda, S., Paschalidis, I. Ch., Vakili, P., and Kozakov, D. 2015. Energy minimization on manifolds for docking flexible molecules. Journal of Chemical Theory and Computation, 11: 1063–1076.
Modis, Y., Ogata, S., Clements, D., and Harrison, S.C. 2003. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proceedings of the National Academy of Sciences, 100: 6986–6991.
Modis, Yorgo, Ogata, S., Clements, D., and Harrison, S.C. 2004. Structure of the dengue virus envelope protein after membrane fusion. Nature, 427: 313–319.
Modis, Yorgo, Ogata, S., Clements, D., and Harrison, S.C. 2005. Variable Surface Epitopes in the Crystal Structure of Dengue Virus Type 3 Envelope Glycoprotein. Journal of Virology, 79: 1223–1231.
Moghadamtousi, S., Nikzad, S., Kadir, H., Abubakar, S., and Zandi, K. 2015. Potential antiviral agents from Marine Fungi: An Overview. Marine Drugs, 13: 4520–4538.
Munda, S., Saikia, P., and Lal, M. 2018. Chemical composition and biological activity of essential oil of Kaempferia galanga: A review. Journal of Essential Oil Research, 30: 303–308.
Murcko, M.A. and Ajay. 1995. Computational methods to predict binding free energy in ligand-receptor complexes. Journal of Medicinal Chemistry, 38: 4953–4967.
Norazharuddin, H. and Lai, N.S. 2018. Roles and prospects of dengue virus non-structural proteins as antiviral targets: an easy digest. The Malaysian Journal of Medical Sciences : MJMS, 25: 6–15.
Olagnier, D., Peri, S., Steel, C., van Montfoort, N., Chiang, C., Beljanski, V., Slifker, M., He, Z., Nichols, C.N., and Lin, R. 2014. Cellular oxidative stress response controls the antiviral and apoptotic programs in dengue virus-infected dendritic cells. PLoS Pathog 10: e1004566.
Pantsar, T. and Poso, A. 2018. Binding affinity via docking: fact and fiction. Molecules : A Journal of Synthetic Chemistry and Natural Product Chemistry, 23.
Parwata, I.M.O.A. 2016. Diktat obat tradisional. Universitas Udayana.
Rouvinski, A., Dejnirattisai, W., Guardado-Calvo, P., Vaney, M.-C., Sharma, A., Duquerroy, S., Supasa, P., Wongwiwat, W., Haouz, A., Barba-Spaeth, G., Mongkolsapaya, J., Rey, F.A., and Screaton, G.R. 2017. Covalently linked dengue virus envelope glycoprotein dimers reduce exposure of the immunodominant fusion loop epitope. Nature Communications, 8: 15411.
Salim, Z. and Munadi, E. 2017. Info Komoditi Tanaman Obat. Badan Pengkajian dan Pengembangan Perdagangan Kementerian Perdagangan Republik Indonesia.
Sarwar, M.W., Riaz, A., Dilshad, S.M.R., Al-Qahtani, A., Nawaz-Ul-Rehman, M.S, and Mubin, M. 2018. Structure activity relationship (SAR) and quantitative structure activity relationship (QSAR) studies showed plant flavonoids as potential inhibitors of dengue NS2B-NS3 protease. BMC Structural Biology, 18: 6.
Shah, M., Wadood, A., Rahman, Z., and Husnain, T. 2013. Interaction and inhibition of dengue envelope glycoprotein with mammalian receptor DC-Sign, an in-silico approach. PLoS ONE, 8: e59211.
Shang, A., Cao, S.-Y., Xu, X.-Y., Gan, R.-Y., Tang, G.-Y., Corke, H., Mavumengwana, V., and Li, H.-B. 2019. Bioactive compounds and biological functions of garlic (Allium sativum L.). Foods, 8: 246.
Silva, E.M., Conde, J.N., Allonso, D., Ventura, G.T., Coelho, D.R., Carneiro, P.H., Silva, M.L., Paes, M.V., Rabelo, K., Weissmuller, G., et al. 2019. Dengue virus nonstructural 3 protein interacts directly with human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and reduces its glycolytic activity. Scientific Reports, 9: 2651.
Siswanto. 2012. Saintifikasi jamu sebagai upaya terobosan untuk mendapatkan bukti ilmiah tentang manfaat dan keamanan jamu. Buletin Penelitian Sistem Kesehatan, 15: 203–211.
Smit, J. M., Moesker, B., Rodenhuis-Zybert, I., and Wilschut, J. 2011. Flavivirus cell entry and membrane fusion. Viruses, 3: 160–171.
Sousounis, K., Haney, C.E., Cao, J., Sunchu, B., and Tsonis, P.A. 2012. Conservation of the three-dimensional structure in non-homologous or unrelated proteins. Human Genomics, 6: 10.
Subositi, D. and Wahyono, S. 2019. Study of the genus Curcuma in Indonesia used as traditional herbal medicines. Biodiversitas Journal of Biological Diversity, 20.
Sugita, P., Octaviana, N., Wukirsari, T., and Rahayu, D.U.C. 2018. Chemical constituent and antioxidant activity of methanol extract from Indonesian Curcuma aeruginosa roxb. Rhizome. Journal of Pharmacy Research, 12: 5.
Sumayyah, S. and Salsabila, N. 2017. Obat tradisional: antara khasiat dan efek sampingnya. Majalah Farmasetika, 2: 1–4.
Tahir, R.A., Wu, H., Rizwan, M.A., Jafar, T.H., Saleem, S., and Sehgal, S.A. 2018. Immunoinformatics and molecular docking studies reveal potential epitope-based peptide vaccine against DENV-NS3 protein. Journal of Theoretical Biology, 459: 162–170.
Theanphong, O., Mingvanish, W., and Kirdmanee, C. 2015. Chemical constituents and biological activities of essential oil from Curcuma aeruginosa roxb. Rhizome. 12.
Tian, Y.-S., Zhou, Y., Takagi, T., Kameoka, M., and Kawashita, N. 2018. Dengue virus and its inhibitors: a brief review. Chemical and Pharmaceutical Bulletin, 66: 191–206.
Trott, O. and Olson, A.J. 2009. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, NA-NA.
Vankadari, N. 2020. Arbidol: A potential antiviral drug for the treatment of SARS-CoV-2 by blocking trimerization of the spike glycoprotein. International Journal of Antimicrobial Agents, 56: 105998.
Wallace, A.C., Laskowski, R.A., and Thornton, J.M. 1995. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Engineering, 8: 127–134.
Wang, X., Cao, R., Zhang, H., Liu, J., Xu, M., Hu, H., Li, Y., Zhao, L., Li, W., Sun, X., et al. 2020. The anti-influenza virus drug, Arbidol is an efficient inhibitor of SARS-CoV-2 in vitro. Cell Discovery, 6: 1–5.
Wang, T-Y., Li, Q., and Bi, K-S. 2018. Bioactive flavonoids in medicinal plants: Structure, activity and biological fate. Asian Journal of Pharmaceutical Sciences, 13: 12-23.
Warkentien, T. 2016. Dengue Fever: Historical Perspective and the Global Response. Journal of Infectious Diseases and Epidemiology, 2.
Woerdenbag, H.J. and Kayser, O. 2014. Jamu: Indonesian traditional herbal medicine towards rational phytopharmacological use. Journal of Herbal Medicine, 4: 51–73.
World Health Organization & Department of Control of Neglected Tropical Diseases. 2017. Integrating neglected tropical diseases into global health and development: Fourth WHO report on neglected tropical diseases. World Health Organization.
Yan, W., Ying, W., ZhiHua, L., ChengFang, W., JianYu, W., XiaoLan, L., PingJuan, W., ZhaoFeng, Z., ShuShan, D., DongYe, H. et al. 2014. Composition of the essential oil from Alpinia galanga rhizomes and its bioactivity on Lasioderma serricorne. Bulletin of Insectology, 67: 247–254.
Zou, M., Liu, H., Li, J., Yao, X., Chen, Y., Ke, C., and Liu, S. 2020. Structure-activity relationship of flavonoid bifunctional inhibitors against Zika virus infection. Biochemical Pharmacology, 177: 113962.
OPEN access freely available online
Chiang Mai University Journal of Natural Sciences [ISSN 16851994]
Chiang Mai University, Thailand
https://cmuj.cmu.ac.th
Arief Hidayatullah1, Wira Eka Putra1,2,*, Sustiprijatno3, Galuh Wening Permatasari4, Wa Ode Salma5, Diana Widiastuti6, Hendra Susanto1,2, Bayyinatul Muchtaromah7, Dewi Ratih Tirto Sari8,9, Febby Nurdiya Ningsih10, Muhammad Fikri Heikal1, Alyana Mahdavikia Rosyada Yusuf1, and Aliyya Suci Arizona1
1 Department of Biology, Faculty of Mathematics and Natural Sciences, Universitas Negeri Malang, East Java, Indonesia
2 Department of Biotechnology, Faculty of Mathematics and Natural Sciences, Universitas Negeri Malang, East Java, Indonesia
3 Indonesian Center for Agricultural Biotechnology and Genetic Resources Research and Development, West Java, Indonesia
4 Indonesian Research Institute for Biotechnology and Bioindustry, Bogor, West Java, Indonesia
5 Department of Nutrition, Faculty of Public Health, Halu Oleo University, Indonesia
6 Department of Chemistry, Faculty of Mathematics and Natural Science, Universitas Pakuan, West Java, Indonesia
7 Department of Biology, Faculty of Science and Technology, Universitas Islam Negeri Maulana Malik Ibrahim, East Java, Indonesia
8 Department of Biology, Faculty of Mathematics and Natural Sciences, Brawijaya University, Indonesia
9 Research center of Smart Molecule of Natural Genetics Resources, Brawijaya University, Indonesia
10 Center of Pharmaceutical and Medical Technology, Agency for the Assessment and Application of Technology (BPPT), Serpong, South Tangerang, Indonesia.
Corresponding author: Wira Eka Putra, E-mail: wira.putra.fmipa@um.ac.id
Total Article Views
Editor: Korakot Nganvongpanit, Chiang Mai University, Thailand
Article history:
Received: January 6, 2021;
Revised: April 17, 2021;
Accepted: April 27, 2021;
Published online: May 13, 2021

