
Discovery of Drug Candidate from Various Natural Products as Potential Novel Dengue Virus Nonstructural Protein 5 (NS5) Inhibitor
Arief Hidayatullah, Wira Eka Putra* , Wa Ode Salma, Bayyinatul Muchtaromah, Galuh Wening Permatasari, Hendra Susanto, Diana Widiastuti, and Muhammad KismurtonoPublished Date : 2020-12-17
DOI : https://doi.org/10.12982/CMUJNS.2021.018
Journal Issues : Number 1, January-March 2021
Abstract Dengue is a severe health problem, especially in under-developing countries like Indonesia. The government and the community have made various efforts. However, the current programs are focusing on controlling disease vector. Medical treatment so far only treats the symptoms. On the other hand, the development of antiviral compounds has been dominated by synthetic compounds which may have serious side effects. The potential of natural compounds in Asia is still largely unexplored. Dengue virus nonstructural protein 5 (NS5) is highly conserved in all serotypes. This study aims to screen several potential natural compounds to be developed as specific antiviral compounds targeting DENV NS5. This research conducted by using in silico approaches including data mining, molecular docking, and 2D-3D visualization. About 170 compounds from 88 organisms analyzed to get the top nine compounds with the highest affinity against NS5 protein. The top nine compounds are from the halichondramide, trigocherriolide, quinadoline, tryptoquivaline, and zingiberene groups. The docking results showed that these potential compounds have an affinity of about 67-100% higher than ribavirin. The visualization results show that these compounds are grouped into three different binding sites. They are RdRP, MTase, and the junction. Compounds that bind in RdRP are thought to interfere with the process of RNA viral synthesis, compounds that bind to MTase are thought to disrupt the viral RNA capping process, while compounds that bind between the two domains are thought to interfere NS5 structural integrity. The compound with the highest affinity is halishigamide A. Halishigamide A is known to have antimicrobial properties, but the action mechanism is still unknown. Compared to the control drug, the halishigamide A tends to have more dynamic movement and interaction to the target protein’s binding site due to its residues’ chemicals motive. Together, these results showed the potency of natural products as specific antiviral compounds targeting DENV NS5.
Keywords: Antiviral compounds, Bioinformatics, DENV-2 NS5, Halishigamide A, Ribavirin
Funding: This study is funded by PNBP Fakultas Matematika dan Ilmu Pengetahuan Alam, Universitas Negeri Malang research grant (Wira Eka Putra).
Citation: Hidayatullah, A., Putra, W.E., Salma, W.O., Muchtaromah, B., Permatasari, G.W., Susanto, H., Widiastuti, D., and Kismurtono, M. 2021. Discovery of drug candidate from various natural products as potential novel dengue virus nonstructural protein 5 (ns5) inhibitor. CMUJ. Nat. Sci. 20(1): e2021018.
INTRODUCTION
Dengue generally caused by infection of four types of dengue virus (DENV): DENV-1, -2, -3, and -4, which transmitted through mosquito bites from the genus Aedes, especially Aedes aegepti and Aedes albopticus. This entity is one of the most common and dangerous mosquito-borne diseases in the world today because cases of dengue fever and dengue hemorrhagic fever can occur undoubtedly in almost all over the world (Warkentien, 2016). WHO classifies this disease as a neglected tropical disease because it is generally endemic in some parts of Asia-Pacific and Africa, which incidentally considered as an under-developing region. At least 2.5 billion people worldwide have at risk of dengue, with an average annual case of 50 million cases spread in more than 100 countries around the world. Every year about 500,000 peoples must be hospitalized and, among them, 20,000 death (Warkentien, 2016; World Health Organization & Department of Control of Neglected Tropical Diseases, 2017). Globally, the tendency of infection every year even shows an increasing trend, at least in the last 30 years. The actual infection rate is likely to be higher than estimated by five times as there are still many unreported cases, especially from areas with inadequate health facilities (Bhatt et al., 2013).
Indonesia is one of the most severely affected countries by dengue in the world (Nadjib et al., 2019). Indonesia is a tropical country that is certainly very suitable for the development of vector diseases such as mosquitoes. Besides, the rapidly developing urban and suburban areas have also made the disease more widespread (Haryanto, 2018). Also, factors that influence the dengue distribution in Indonesia are people mobility and the flow of foreign tourists, especially in well-known tourist destinations such as Bali, Yogyakarta, and Jakarta (Masyeni et al., 2019). Since it first appeared in 1968, dengue has spread to all provinces in Indonesia.
All DENV serotypes are found in Indonesia. However, the dominant cases are generally caused by DENV-2 and DENV-3 infections because both viruses are endemic in tropical regions such as Indonesia, and the main vector is Aedes aegepti, which is also endemic in the tropics. (Karyanti & Hadinegoro, 2016; Masyeni et al., 2019). However, from year to year, the ratio of mortality due to dengue infection continues to decrease from 41% at the beginning of its appearance, now around 1% of the total cases (Karyanti & Hadinegoro, 2016). Although it looks like a relief, this number is not a sign that currently, dengue is not dangerous because the relatively low percentage of mortality still accompanied by a trend in the incidence of infection that is increasing every year.
The Indonesian government already has a national dengue prevention program. This program emphasizes vector control so that the vector cannot develop in the area where people live. Various efforts such as education for a healthy lifestyle, training for village officials, and the community and research steps have also been undertaken (Sulistyawati et al., 2019). Nevertheless, Indonesia is a developing country that is geographically so widespread that such a program is still yet to be realized evenly throughout Indonesia. Additionally, limited health facilities, research facilities, and human resources hamper the program (Walsham, 2020). Such vector control efforts are carried out because, globally, there are still no vaccines or drugs that are clinically proven to prevent and treat dengue. The current available treatments in hospitals generally only treat or alleviate the symptoms so that the patient's natural immune system will strengthen and fight the dengue infection (Haryanto, 2018). Besides, the treatment ensures that complications do not appear to be more severe in the patients (Rajapakse, 2011).
Some drugs are known to be able to overcome viral infections by acting as an antivirus. One well-known antiviral drug is arbidol made in the Soviet Union, which commonly used to treat influenza, and it is widespread in Russia (Leneva et al., 2009). Most of these antiviral drugs derived from synthetic chemical compounds, only 18 out of 110 antiviral drug products tested from 1981 to 2010, are derivatives of natural compounds such as laninamivir and oseltamivir and zanamivir which are effective against Enterovirus-71 infection. However, over time, more and more synthetic drugs are produced from the modification of organic compounds with structures and properties that are almost similar to organic compounds (Moghadamtousi et al., 2015). Interestingly, many new antiviral compounds show a high level of effectiveness in some viruses. Therefore, the opportunity for new antiviral compounds, especially those which are derivatives of natural materials that have high efficacy in DENV up until the cellular level, encouraging the authors to conduct this research.
Research with similar subjects targeting DENV is rarely conducted. Most of the research aimed at developing DENV antivirus uses synthetic compounds, resulting from development in the laboratory. To this day, there is still no clinically proven and recognized type of antivirus that can overcome or even prevent DENV infection, both from natural compounds and synthetic compounds developed in the laboratory. Therefore, exploration opportunities in this field of research are still very wide open because DENV infection is one of the health problems that are a priority of WHO. One study that sought to explore natural compounds that were considered potential to be developed into antiviral compounds for DENV was conducted by researchers from Kaohsiung Medical University using isolates of several types of carotenoids originating from Palythoa mutuki. That research showed good initial results that able to inhibit the stage of DENV infection but still needed further development to be widely applied
(Lee et al., 2016). Another study conducted in 2013 using flavonoid data isolated from Carica papaya leaf extract in silico showed positive results (Senthilvel et al., 2013). However, there is still no follow up in the form of development of research results like this to be implemented on a larger scale to overcome or even prevent DENV infection.
MATERIALS AND METHODS
Materials
The material used in this study is the structure of natural chemical compounds that we suspect could be used as anti-dengue from various literatures (Andjelic et al., 2008; Atun, 2009; Chung, 2009; Karyawati, 2011; Allard et al., 2012; Moghadamtousi et al., 2015; Apriyanto et al., 2016; Farhaty and Muchtaridi, 2016; Herman, 2019; Koswandy and Ramadhania, 2016; Mentari and Wijayanti, 2016; Zahra and Iskandar, 2017; Dewi and Subarnas, 2018; Balafif et al., 2019;). The structure of compounds is in the form of two-dimensional or three-dimensional structures in the SDF format. We use PubChem (https://pubchem.ncbi.nlm.nih.gov/) as the primary database, with references from credible literature. Each compound's PubChem ID recorded for the validation process.
Preliminary test
The compounds downloaded from the PubChem database then tested whether they meet the Lipinski rule of five or not. This test carried out on the Supercomputing Facility for Bioinformatics and Computational Biology website, IIT Delhi (http://www.scfbio-iitd.res.in/software/drugdesign/lipinski.jsp). Compounds to be tested further must have met these five requirements because this test aims to check whether a compound with specific pharmacological properties has chemical and physical properties that make it possible to use as a drug, especially for human consumption by oral (Lipinski, 2004).
Key protein target
Based on previous literature studies, among all of the non-structural proteins produced in DENV infection, the protein used as the target in this study was DENV2 NS5 (PDB ID: 5ZQK). The selection of this protein-based on this protein's function is vital in the process of virus replication in host cells, mainly responsible for the operation of new viral RNA synthesis. Also, structurally, this protein is relatively conserved in all DENV serotypes so that the hope of this research application can be more comprehensive (Oliveira et al., 2014). The two primary databases for the DENV2 NS5 are RSCB PDB (https://www.rcsb.org/) and UniProt (https://www.uniprot.org/). The data from the two databases are the 3D structure of the target protein in the form of PDB format. The protein data that we got from databases still contain ligands such as S-adenosylmethionine (SAM), glycerol, zinc ion, 1,2-ethanediol, magnesium ion, and also water. It is necessary to purify the protein by removing all the ligands and water attached to the protein for more accurate analysis. This process is through PyMol (https://pymol.org/2/).
Docking process
The docking process is using PyRx (https://pyrx.sourceforge.io/) (Trott & Olson, 2009). The DENV2 NS5 must be converted to the Autodock macromolecular format (pdbqt) before tested. All of the compounds' binding energy must be minimized and then converted to the Autodock macromolecular format (pdbqt) before tested. We use ribavirin (CID: 37542) as the control compound because of its frequent use as an antiviral compound, which has been tested for DENV NS5 (García et al., 2017). The testing process is using a scenario where each protein tested with all compounds that met the Lipinski rule of five. The DENV2 NS5 (5ZQK) tested three times to minimize the appearance of abnormal data. Furthermore, in this present study, grid size used was X: 33.5796, Y: -11.639, and Z: 26.946 and coordinate/ dimensions (Angstrom) used was X: 90.9434, Y: 77.8573, and Z: 90.8368.
Visualization
The docking results visualized with PyMol to see how the bond between the target protein and each compound tested. The docking results in the form of a protein-ligand complex were then analyzed by LigPlot+ (https://www.ebi.ac.uk/thornton-srv/
software/LigPlus/) to find out the systematic bonding between proteins and ligands in the 2D scheme. Out of 170 compounds that we test, we only put the top nine compounds that have the best docking result and have higher affinity than the control. All of them must be natural compounds such as from part of plants, animals, or microbes' metabolites.
RESULTS
Docking result of the top nine selected compounds against DENV2 NS5
Based on the results of verification using Lipinski, out of 203 compounds that have the potential to be antiviral to overcome DENV infection. About 170 compounds met Lipinski criteria, while 53 others did not. Of the 170 compounds tested, they have selected nine compounds to have higher affinity value than the control compound, which is Ribavirin (CID 37542). Compounds selected must be a natural compound derived from an organism in the form of metabolites or deposit in an organism. The docking results are shown in the following table 1.
Table 1. Compounds docking result and 2D plotting for DENV2 NS5 on the A binding site.
|
No. |
Compound |
Source |
ΔG Average |
Amino Acids Residue |
Interaction |
|
1. |
Halishigamide A (CID: 44566626) |
Halichondria sp |
-13.6 kcal/mol |
Tyr630(A), Asp686(A) Ser684(A), Ser733(A) Ser819(A), Cys732(A) Arg760(A), Thr817(A) Thr816(A), Arg752(A) Lys483(A), Lys480(A) Glu482(A), Asn633(A) Gly685(A) |
Hydrophobic contact |
|
2. |
Trigocherriolide C (CID: 71530651) |
Trigonostemon cherrieri (leaves) |
-11.37 kcal/mol |
Lys712(A), Asp557(A), Lys484(A), Leu485(A), Arg481(A), Gln765(A), Met366(A), Tyr781(A), Thr816(A), Trp818(A), Glu482(A), Arg815(A), Thr817(A), Ser819(A), Asp686(A), Asp687(A) |
Hydrophobic contact |
|
|
|
|
|
Lys483(A) |
Hydrophobic contact |
|
|
|
|
|
|
Hydrogen bond (3.20) |
|
|
|
|
|
Arg760(A) |
Hydrophobic contact |
|
|
|
|
|
|
Hydrogen bond (3.28) |
|
|
|
|
|
|
Hydrogen bond (2.87) |
|
|
|
|
|
Ser733(A) |
Hydrophobic contact |
|
|
|
|
|
|
Hydrogen bond (3.03) |
|
3. |
Quinadoline B(CID: 72547182) |
Aspergillus sp |
-11.2 kcal/mol |
Asp562(A), Asp686(A), Ile820(A), Ser684(A), Tyr630(A), Gly685(A), Gln626(A), Lys480(A), Ile497(A), Trp498(A), Ala496(A) |
Hydrophobic contact |
|
|
|
|
|
Arg495(A) |
Hydrophobic contact |
|
|
|
|
|
|
Hydrogen bond (3.08) |
|
4. |
Ribavirin (CID: 37542) |
Antiviral drug (control) |
-6.8 kcal/mol |
Thr816(A) |
Hydrophobic contact |
|
|
|
|
|
Thr817(A) |
Hydrophobic contact |
|
|
|
|
|
|
Hydrogen bond (2.77) |
|
|
|
|
|
Trp818(A) |
Hydrophobic contact |
|
|
|
|
|
|
Hydrogen bond (3.32) |
|
|
|
|
|
Arg752(A) |
Hydrophobic contact |
|
|
|
|
|
|
Hydrogen bond (3.15) |
|
|
|
|
|
Ser733(A) |
Hydrophobic contact |
|
|
|
|
|
|
Hydrogen bond (3.17) |
|
|
|
|
|
Lys483(A) |
Hydrophobic contact |
|
|
|
|
|
|
Hydrogen bond (3.02) |
|
|
|
|
|
Arg760(A) |
Hydrophobic contact |
|
|
|
|
|
|
Hydrogen bond (3.13) |
|
|
|
|
|
|
Hydrogen bond (2.79) |
|
|
|
|
|
Tyr781(A) |
Hydrophobic contact |
|
|
|
|
|
|
Hydrogen bond (3.10) |
|
|
|
|
|
Arg815(A) |
Hydrophobic contact |
|
|
|
|
|
|
Hydrogen bond (3.26) |
|
|
|
|
|
|
|
Note: *Bold indicates the same amino acid as that interact with halishigamide A and Ribavirin. **Underline indicates the same amino acid interacts with trigocherriolide C and Ribavirin
We took nine natural compounds with the greatest affinity value for NS5 including Halishigamide A (-13.6), Trigocherriolide C (-11.37), Quinadoline B (-11.2), Zingiberene (-10.67), Trigocherriolide A (-10.17), Tryptoquivaline F (-10.1), Tryptoquivaline H (-10.1), Quinadoline A (-10.1), and Tryptoquivaline E (-10.1). The docking results showed that the compound with the highest affinity value was halishigamide A, which is a metabolite of the marine sponge of the genus Halichondria. Interestingly, from the top nine compounds, only halishigamide A derived from animal metabolites. While the other eight are metabolites produced by plants or fungus. Halishigamide A affinity value is precisely two times higher when compared to control compounds. The affinity value of these nine compounds ranges from 48-100% higher when compared to control compounds. This result is auspicious because theoretically, the nine compounds tend to bind and interfere with the work of NS5 better when compared to the control.
3D visualization result of the top nine selected compounds against DENV2 NS5
Analysis of 3D visualization results showed that nine compounds have affinity value most to the NS5, and control compounds fall into three main clusters binding site. Binding site A is a binding site where the control compound binds to the NS5 protein, and halishigamide A, trigocherriolide C, and quinadoline B compounds based on docking results are potential compounds with the highest affinity value for NS5. Binding site A, located in a deeper position compared to other binding sites where B and C tend to be outside and not protected by the NS5 protein structure (Figure 1).
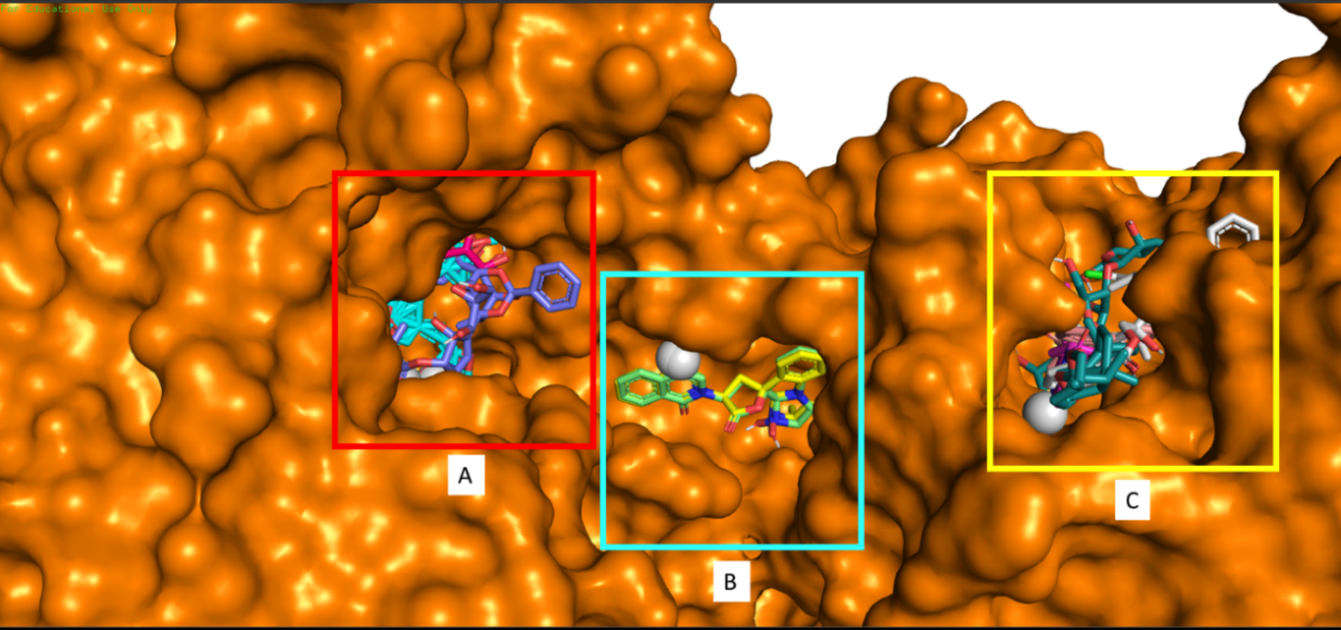
Figure 1. The 3D visualization results show that the tested compounds occupy three main binding sites on DENV2 NS5. Binding site A is a binding site for halishigamide A, trigocherriolide C, quinadoline B, and ribavirin (control). Binding site B is a binding site for tryptoquivaline H, and E. Binding site C is a binding site for tryptoquivaline F, zingiberene, quinadoline A, and trigocherriolide A.
Based on the results (Table 1), about six of nine amino acids (marked in bold) that binds to ribavirin also binds to halishigamide A, and seven of nine amino acids (mark in underline) binds to ribavirin also binds to trigocherriolide C. This means that at least more than 67% of the amino acids bind to ribavirin also binds to these two potential compounds. Hence, it suspected that these two compounds would work similarly to ribavirin.
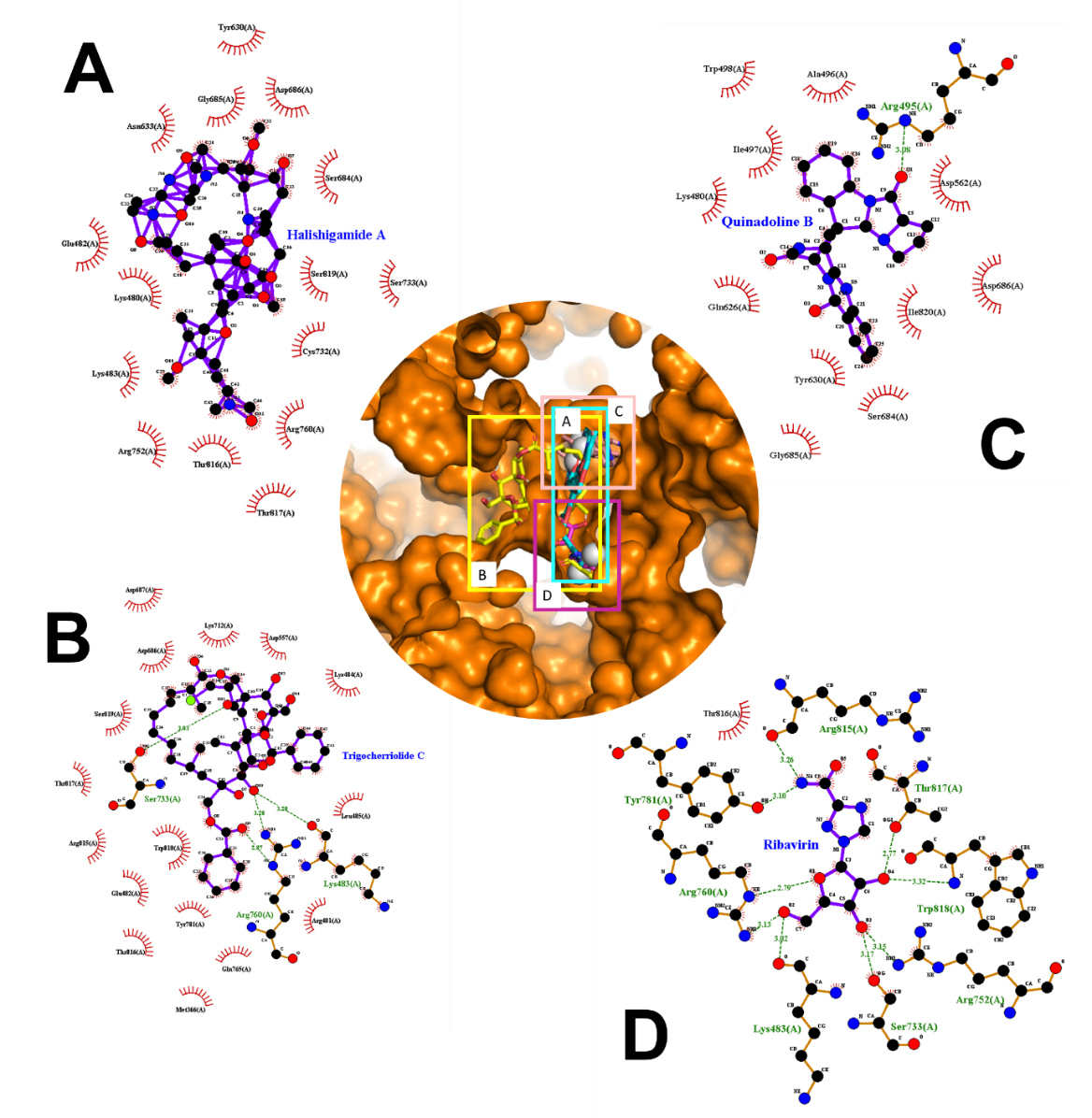
Figure 2. Description of the compounds that bind to the binding site A in 2D and 3D structures. (A) halishigamide A, (B) trigocherriolide C, (C) quinadoline B, and (D) ribavirin as control.
Accordingly, two potential compounds have binding sites that intersect with ribavirin; the two compounds are halishigamide A and trigocherriolide C, which are in compounds with the first and second-largest affinity when referring to the docking results that have done before (Figure 2). This result shows this compound's potential as a substitution compound for ribavirin because it has at least a crossing binding site.
DISCUSSIONS
DENV RNA encodes several types of nonstructural proteins. Generally, these nonstructural proteins play a role in inhibiting the process of host cell immune response. One of the nonstructural proteins that play an essential role in DENV infection is NS5. NS5 is the most significant nonstructural protein produced during an infection with a molecular weight of around 104-105 kDa. Among all nonstructural proteins produced during infection, NS5 is the protein that has the most conserved sequence in all DENV serotypes among seven nonstructural proteins with a ratio of 70 % identic protein sequences in all serotypes. The NS5 sequences conserved among all serotypes of DENV make it ideal as a target for antiviral compounds. It because it can provide a broad spectrum of antiviral compounds that are not only effective in overcoming one DENV serotype but have the opportunity to be applied to all DENV serotypes (El Sahili & Lescar, 2017). Conserved NS5 sequences also found in other viruses originating from the Flaviviridae family such as ZIKV, yellow fever virus, and Japanese encephalitis virus. So that the potential application of potential compounds as antivirals targeting specific proteins such as NS5 is not only limited to DENV but also other viruses originating from one family (Dubankova & Boura, 2019). NS5, along with NS3, is an essential protein that plays a significant role in the DENV replication process (Figure 3).
DENV NS5 consists of two main domains. They are a methyltransferase domain (MTase) at its N-terminal end and an RNA-dependent RNA polymerase (RdRP) at its C-terminus. The MTase domain functions for the synthesis of guanylyltransferase, guanine-N7-methyltransferase, and nucleoside-2'O-methyltransferase, which play a role in the viral RNA capping process, while the RdRP domain plays a vital role in the process of synthesizing new RNA (Klema et al., 2016). Specific domain RdRP activity synthesizes viral RNA, making it a potential marker of silencing by potential antiviral compounds. This characteristic is due to the NS5 domain RdRP activity is exclusively found in cases of viral infection and never found in the host cell itself. This domain conserved throughout the family Flaviviridae serotypes even for the most critical role of RNA viral replication (Ashour et al., 2009; El Sahili & Lescar, 2017). Because of this characteristic, NS5 is an immunogenic molecule, which means that the presence of NS5 will trigger an immune response in infected cells. Because of its conservative, it also can be developed as a potential antigen for more accurate dengue infection diagnosis in the future (Zhang et al., 2019).
In previous research, DENV NS5 has identified as being an IFN antagonist that directly or indirectly associates with STAT2, a necessary component of the ISGF3 transcription complex, leading to its proteolytic processing (Figure 3). While the expression of DENV NS5 alone resulted in STAT2 binding, DENV NS5 to target STAT2 for degradation required the presence of a protease cleavage signal upstream of the N terminus of NS5. With the subsequent coexpression of this precursor form of NS5 with the relevant protease to target the cleavage site, mirroring the NS5 processing occurring in the context of the DENV polyprotein (Ashour et al., 2009). Thus it is known that in addition to having a vital function for viral RNA replication, NS5 generated by DENV also serves to inhibit the innate immune response in host cells.
Some studies on potential compounds as DENV NS5 inhibitors that had previously done generally using candidate compounds a synthesis that developed in the laboratory. One example is RK-0404678, which tested on the RdRP domain to demonstrate the potential of these compounds as the backbone in the development of antiviral compounds targeting the DENV NS5 (Shimizu et al., 2019). Other compounds such as ZINC64717952 and ZINC39563464 also reported promising results when tested using in silico methods at NS5 ZIKV. ZIKV still considered as one family with DENV (Ramharack & Soliman, 2018). Some compounds and drugs that have used as inhibitors for NS5 also did not show promising results. One of them is sinefungin, which tested as an NS5 inhibitor, and the results show that sinefungin has a six times higher affinity than S-adenosyl methionine, better known as SAM.
Nevertheless, the greatest weakness of sinefungin is that it is difficult for these compounds to enter the cell, and further studies do not show promising development. S-adenosyl homocysteine has also tested as an NS5 inhibitor, but the results obtained are also not very promising, especially because this compound is difficult to enter the cell. GMP also produces almost identical results. Drugs such as ribavirin, which commonly used as a benchmark in studies like this, are also not very clinically effective in inhibiting NS5, primarily because of its high toxicity. Even drugs such as balapiravir, which assessed as potential antiviral drugs for hepatitis C, did not show promising results in clinical trials (García et al., 2017). Ribavirin has often been used as a benchmark to measure the effectiveness of a compound against DENV NS5. In this study, we used ribavirin as a control.
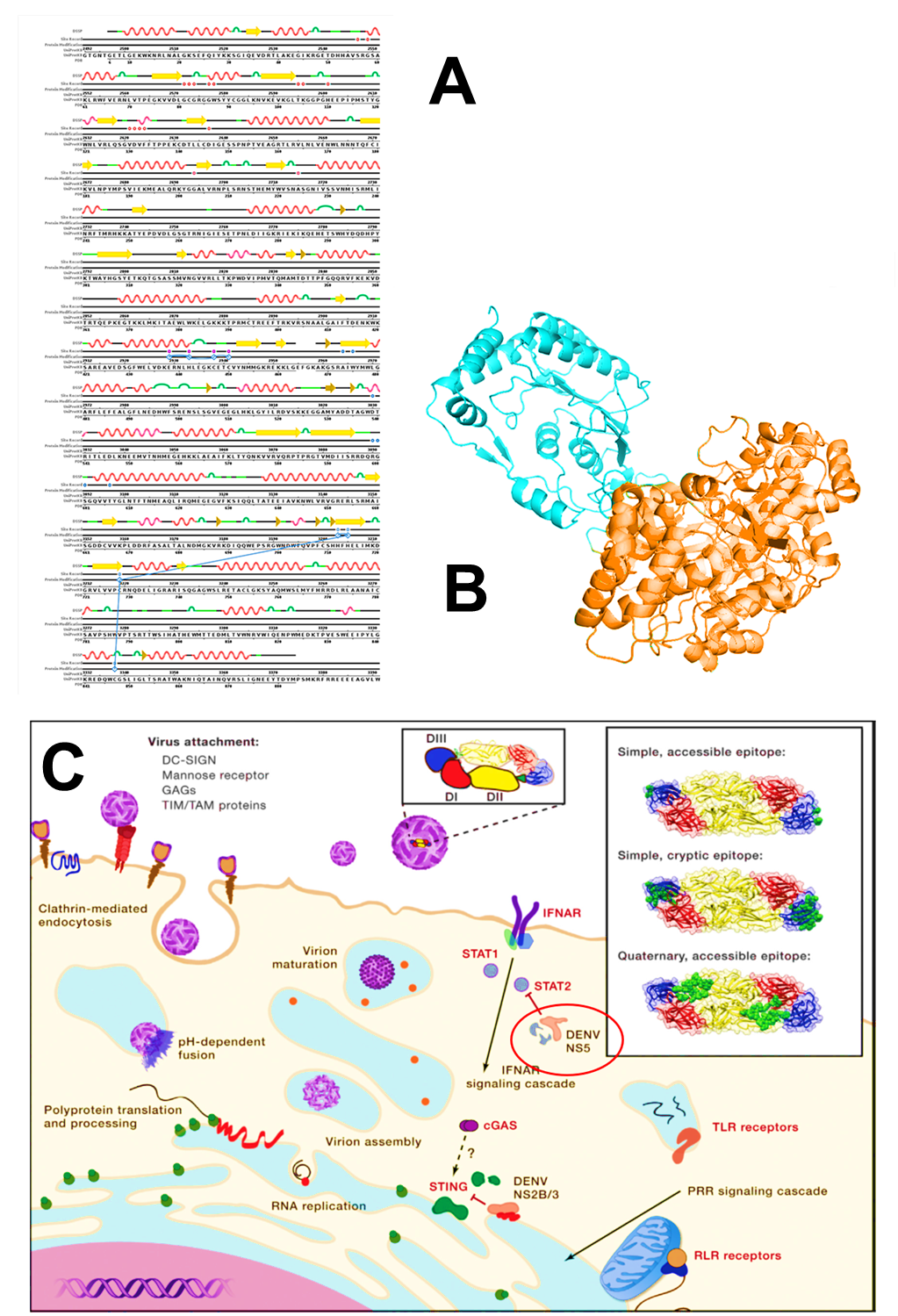
Figure 3. (A) Full-length sequence from DENV2 NS5 (PDB ID: 5ZQK) (B) 3D visualization of DENV2 NS5 results. In this visualization, two domains appear. The cyan colored domain is MTase, while the orange domain is RdRP (Sehnal et al., 2018). (C) DENV infection mechanism at the cellular level. DENV infection begins with the process of endocytosis. Endocytosis starts with the bond between the viral membrane proteins with several kinds of host cell membrane proteins such as DC-SIGN, mannose receptor, GAGs, and TIM/TAM. After endocytosis takes place, the DENV replication process begins. The host cell has an immune response mechanism, but DENV will inhibit the immune response to the nonstructural protein synthesis. DENV has evolved mechanisms, including the NS2B/3-mediated cleavage of STING and NS5-mediated degradation of STAT2, to antagonize antiviral pathways (Diamond and Pierson, 2015).
The docking results showed several potential compounds that at least had a higher affinity for DENV2 NS5 when compared to ribavirin. In these studies, a compound with the highest affinity, even double the affinity of Ribavirin to NS5 is halishigamide A. Halishigamide A is a compound from the class of macrolides or halichondramides which is an oxazoles-containing macrolide isolated from marine sponges of the genus Halichondria from the Pacific. This compound was first known as an antibiotic and antifungal compound, which in testing shown to show significant antifungal activity against Candida albicans and Trichophyton mentagrophytes, which were explored at least since 1987 (Tm, 2019). In general, the macrolide is a protein synthesis inhibitor. It will inhibit the translation process in the ribosome by inhibiting the polypeptide chain lengthening process mediated by peptidyltransferase or bind to the 50S ribosomal subunit and targeting the 23S ribosomal RNA molecule. Nevertheless, there is no more detailed explanation about the mechanism of action of the more specific compound, such as halichondramides or halishigamide A (Menninger, 1995). Halishigamide A is the only compound that is a metabolite from animals.
The next compound that is also a potent inhibitor of DENV2 NS5 is from the trigocherriolide group isolated from the leaves of Trigonostemon cherrieri. There are two trigocherriolide compounds which have a high-affinity value for DENV2 NS5. They are trigocherriolide C, and trigocherriolide A. Trigocherriolide itself is part of diterpenoids, a large and structurally diverse class of natural products derived from four C5 isoprene units joined in a head-tail fashion. Both of these trigocherriolide compounds have an affinity value above -10 kcal/mol. Trigocherriolide A has previously been investigated as an antiviral for the chikungunya virus (CHIKV) and has shown quite positive results in inhibiting the CHIKV replication process. On the other hand, other studies show that the Trigocherriolide group also inhibits CHIKV pathway enzymes. When compared with the results of docking on DENV2 NS5, the Trigocherriolide compounds can become potential antivirals for DENV (Bourjot et al., 2014; Jayaprakash et al., 2019). Biochemical studies show that Trigocherriolide, when tested on the CHIKV and NSR D5V RDRP domains, show that trigocherriolide is significantly more potent as an antiviral when compared to chloroquine (Allard et al., 2012).
Another group of compounds that could potentially be anti-DENV quinadoline B and A. Basically, quinadoline is a metabolite isolated from the genus Aspergillus fungi belonging to the group of alkaloid compounds (Hart, 2010). One source stated that the fungus genus Aspergillus which produced this compound, was isolated from the mangrove leaves Sonneratia hainanensis (Guo et al., 2018). One of the most commonly mentioned functions possessed by this type of compound is that it can inhibit the formation of lipid droplets on macrophages to reduce the risk of atherosclerosis (Koyama et al., 2008; Wu and Ma, 2013). Some studies state that quinadoline shows the inhibitory activity of the influenza A (H1N1) virus. However, the researchers did not explain how this group of compounds works to inhibit the influenza A (H1N1) virus (Roy, 2017). One study mentioned that testing quinadoline A on DENV2 showed quite positive results. The results indicated that quinadoline A does not show significant cytotoxicity when tested on cultured cells. The test results showed that the quinadoline group generally showed positive results in inhibiting the process of synthesis of new viruses. There are differences in quinadoline A and B results where quinadoline B shows a more significant inhibitory effect on H1N1 compared to quinadoline A. This research does not explain the mechanism further by which this compound acts as an antiviral agent for DENV. However, the presumption is that this compound inhibits the formation of new viruses and the release of new viruses from host cells (Shang et al., 2018). These results are entirely consistent with the results obtained in this study where quinadoline A and B showed positive results when tested against DENV2 NS5 in silico. Also, the docking results show that the affinity value of quinadoline B is higher than that of quinadoline A.
Zingiberene is a joint compound found in plants from the Zingiberaceae family, such as temulawak (Curcuma zanthorrhiza) and ginger (Zingiber officinale). These volatile compounds generally make essential oils in the rhizomes. Chemically, this compound is a monocyclic sesquiterpene compound, which means it has only one aromatic ring attached to a terpenes structure. Naturally, this compound serves as a mechanism of self-defense plants from the Zingiberaceae family from insect pests (Bone & Mills, 2013). These compounds are generally known as antioxidants and anti-inflammatory.
Some studies suggest that zingiberene also has the effect of antibacterial, antifungal, antiviral even. However, this antiviral claim still requires further evidence and research (Mao et al., 2019). Research on the impact of the use of zingiberene as DENV antiviral even hard to find, so the explanation of the mechanism in detail how these compounds work as antivirus is still minimal. Some studies suggest that ginger extract can inhibit the process of plaque formation in the respiratory tract caused by the infection of the human respiratory syncytial virus (HRSV). It added that ginger extract could inhibit the viral attachment process on HRSV. Also, ginger extract can inhibit the work of the hepatitis C virus. However, it has not yet explained further which compounds of the ginger extract play the most crucial role in inhibiting the activity of the virus and its mechanism of action (Mao et al., 2019). It also has not been found studies that review the effects of inhibition activity against DENV by zingiberene.
The last group of potential compounds as DENV antivirals in this study is tryptoquivaline. This compound is a metabolite compound produced by the fungus Cladosporium sp. In general, these compounds belong to a large group of alkaloids as quinadoline (Moghadamtousi et al., 2015). Some types of tryptoquivaline are known to have antifungal effects on species such as Candida albicans and Fusarium solani (Zhang et al., 2016). The results of previous studies also showed that tryptoquivaline was able to inhibit the activity of the influenza A (H1N1) virus, almost the same as quinadoline, which also showed positive results that were better than ribavirin (Moghadamtousi et al., 2015). Just like in quinadoline, mechanism detail how tryptoquivaline inhibits the activity of H1N1 is also not spelled out clearly in the previous study. Besides, in-depth reviews of the mechanism of these compounds as potential antivirals are also quite uncommon. However, based on docking results that show values above -10, the potential of this compound to be developed into a DENV antivirus targeting NS5 is quite good.
Nine compounds that are considered the most potential as an anti-NS5 in the present study divided into three clusters of binding sites. Binding site A is a binding site located in the middle of the RdRP domain, binding site B is a binding site located between two NS5 domains, and binding site C is a binding site in the middle of the MTase domain. Ribavirin as a control occupies binding site A, which is in the RdRP domain. Ribavirin, known as a broad spectrum antivirus, works as an analog of guanosine. In host cells, ribavirin will convert into ribavirin monophosphate (RMP), then RMP is converted by ITPase to ribavirin triphosphate (RTP). RTP can directly inhibit RdRP in concentrations between 10-100 μM, thereby interfering with forming new viral RNA. RTP will bind to the existing nucleotide-binding site in RNA polymerase so that it acts as a competitive inhibitor of nucleotides that will be linked together to form new viral RNA so that the process of viral RNA elongation disrupted. Viral RNA that created becomes nonfunctional or damaged. At lower concentrations (10 μM), RMP will inhibit the action of inosine monophosphate dehydrogenase, causing GTP depletion and will interfere with the mRNA-capping process. Whereas at high concentrations of more than 100 μM, RTP can cause mutations in the viral genome because of the nature of ribavirin, which works as an analog of guanine, which can form pseudo-base pairs with cytidine and uridine and increase the transition of G-A and A-G to the viral genome. Unfortunately, some reports indicate that the bond between ribavirin and NS5 is unstable, reducing the drug (Nyström et al., 2019). Compounds that have the same binding site as ribavirin are the three compounds with the highest affinity values. Thus, it expected that these compounds can have a mechanism of action and targets similar to ribavirin in inhibiting the synthesis of new RNA but with more reliable because the tested compounds' affinity value is 65-100% higher than ribavirin. We suspect that the compounds tested will act as potent competitive inhibitors of nucleotides, which will assemble into new viral RNA. All three compounds are halishigamide A, trigocherriolide C, and quinadoline B. Based on Table 1, all the residues that interact with the three potential compounds and ribavirin are the residue that is on RdRP domain because it was in the range between residues 272-900 (Potisopon et al., 2014). Ribavirin has the most hydrogen bonds compared to all compounds that have the same binding site even though it has the lowest affinity value compared to other compounds.
On the other hand, halishigamide A has the highest affinity value as high as twice the affinity of ribavirin. Still, all contact with residues is hydrophobic contact, which is incidentally weaker than hydrogen bonds. Trigocherriolide C has an affinity value of
67% higher than ribavirin, but of the 19 contact residues, only 3 of them have hydrogen bonds. Although quinadoline B is in the binding site complex A, just like halishigamide A and ribavirin, quinadoline B has no contact with the same residue as ribavirin as if referring to figure 2 shows that the quinadoline B does not intersect with ribavirin. However, quinadoline B has an affinity value of 64% higher than ribavirin but only has one hydrogen bond, which is far less than ribavirin, which has eight hydrogen bonds.
The second binding site, binding site B, is located at the junction between MTase and RdRP. This area plays a vital role as a determinant of the flavivirus NS5 conformation drawn up by the two domains. This junction structure can affect the orientation of each domain, domain folding process, and the process of catalysis polymerase. Changes or even loss of this part will affect the polymerase activity. This junction maintains a canonical fold of RdRP, which plays a vital role in initiating new viral RNA synthesis in the RdRP domain (Wu et al., 2015). This junction area will improve the initiation process of polymerase work by enhancing the turnover of the RNA and NTP substrates and improve protein thermostability, so it still can be stable at the higher temperature (Lim, Koh, et al., 2013). Thus, this junction is essential for the integrity and performance of NS5, which is also possible as a target domain for antiviral compounds. Antiviral compounds are targeting this junction that expected to affect several factors: the integrity and stability of the NS5 protein itself, flexibility, and orientation of functional domains and the function of the functional domains indirectly. The two compounds that bind to this site are tryptoquivaline H and E, both with an affinity value of -10.1, which is at least 48 % higher when compared to the affinity of ribavirin on the binding site A. Based on the results described in table 1, it shows that tryptoquivaline H and E have hydrophobic contacts with residues in two NS5 domains at the same time which makes it at the junction between the two domains. Tryptoquivaline H and E have the same amount of hydrophobic contact, which is 10. Tryptoquivaline H has contact with residues Pro136 (A), Glu380 (A), Lys492 (A), Lys381 (A), Val377 (A), Arg376 (A), Tyr112 (A), Trp144 (A), Pro138 (A), and Phe372 (A) while tryptoquivaline E has contact with residues Pro136 (A), Lys492 (A), Lys381 (A), Val377 (A), Glu380 (A), Arg376 (A). Lys379 (A), Trp144 (A), Tyr112 (A), and Pro138 (A). In these results, it appears that the residue in contact with tryptoquivaline H and E is 70% identical. Thus, the possibility of substitution between the two compounds is quite large. Pro136 (A), Tyr112 (A), Trp144 (A), and Pro138 (A) residues are residues in the MTase domain (NS5-MTase, residues 1–263, 30 kDa), while Glu380 (A), Lys492 (A), Lys381 (A), Val377 (A), Arg376 (A), Glu380 (A), and Lys379 (A) are residues that are in the RdRP domain (NS5-RdRP, residues 272–900, 74 kDa) (Potisopon et al., 2014). It appears that tryptoquivaline H and E have direct contact with both domains so that they are placed right in the junction area, and the possibility of working as a disturbance agent, structural and functional, is also quite possible. However, the drawback is tryptoquivaline H and E only have hydrophobic contact with the residues, which is weaker than hydrogen bonds.
The third binding site is known as binding site C, located in the MTase domain. Four of the nine compounds tested have binding sites in this domain. These compounds include zingiberene, trigocherriolide A, tryptoquivaline F, and quinadoline A. Among the four compounds that have binding sites at binding sites C have at least the same contact residue of 87%. Based on sequences of residues that interact with these four compounds, all residues are in the MTase domain (residues 52-238). Considering the function of the MTase domain for the synthesis of guanylyltransferase, guanine-N7-methyltransferase, and nucleoside-2'O-methyltransferase which plays a role in the viral RNA capping process, the presence of these compounds is likely to disrupt these processes which will result in damage to RNA resulting from NS5 synthesis so that the RNA will decrease in quality or even become nonfunctional. The MTase domain in DENV2 NS5 generally binds to S-adenosyl methionine (SAM), which acts as a methyl donor in the RNA methylation process (Potisopon et al., 2014). These four compounds have at least contact with the same residue with SAM up to 71% so that the possibility of these four compounds can be a competitive inhibitor of SAM that disrupts the work of MTase because of the lack of the primary methyl donor. Zingiberene has high-affinity values up to -10.67. Moreover, the Zingiberene compound has high-affinity values up to -10.67. Also, eleven out of twenty residues in contact with zingiberene have hydrogen bonds, which are theoretically more robust when compared to hydrophobic contact, so the bond between zingiberene and MTase domain theoretically stronger compared to other compounds.
Halishigamide A is a compound that has identified as a metabolite products marine sponge of the genus Halichondria originating from the Pacific at least since 33 years ago. However, the study on the benefits and its application in human life is not enough yet. Research that has done before generally only mentions macrolide or halishigamide in general, both its chemical nature and structure, as well as some of its functions. Generally, research only mentions that halichondramide, which is halishigamide A, includes inhibitory activity in Candida albicans and Trichophyton mentagrophytes. (Karpiński, 2019). Halishigamide A is the ammonia adduct of halichondramide at C-5 (Matsunaga, 2006). Because halishigamide A is hydrophobic macrolides, the molecules of these compounds will have no difficulty in penetrating the target cell membrane (Bryskier & Bergogne-Bérézin, 2005). There is no further explanation of how it works as an antimicrobial. One study says that halichondramide generally has a low selectivity index that could potentially be a cure for malaria after further development, especially to reduce cytotoxicity (Donia & Hamann, 2003). However, based on an in silico study, halishigamide A occupies the top position as a natural compound with the highest affinity for DENV2 NS5. The most striking difference between halishigamide A and ribavirin as a control is the bond with NS5. In terms of amount, halishigamide A has 67% more contact with NS5 than ribavirin. Also, halishigamide A and ribavirin have contact with the same six residues. They are Ser733 (A), Arg760 (A), Thr817 (A), Thr816 (A), Arg752 (A), and Lys483 (A). However, the bond between the residue and the two compounds is not identical. Only the contact between Thr817 (A), and Thr816 (A) are completely identical in both compounds, that is, both residues have hydrophobic contact with both compounds. Whereas in Ser733 (A), Arg760 (A), Arg752 (A), and Lys483 (A), there is a difference between the two compounds. In halishigamide A, all of the residues only have hydrophobic contact, but in ribavirin, all the residues have hydrophobic contact and hydrogen bonding except Thr816 (A) that only has hydrophobic contact. This difference causes the difference in bond energy between the two compounds to NS5 so that the conformation can determine the most stable molecular geometry. In halishigamide A, all residues that have hydrophobic contact with this compound are hydrophilic residues, except for Tyr630 (A), a hydrophobic residue. In ribavirin, seven out of nine binding residues are hydrophilic residues. All of these residues have hydrogen bonds with the compound except for Thr816 (A). Meanwhile, Trp818 (A) and Tyr781 (A) are hydrophobic residues, and both have hydrophobic contact and hydrogen bond. With a much higher affinity and a more potent molecule than ribavirin, halishigamide A has the potential to be developed into an anti-dengue, explicitly targeting the RdRP domain in NS5.
This study only briefly discusses nine natural compounds in silico tests that have the best results among the 170 tested natural compounds derived from 88 different organisms. Due to our limitations, we cannot test all the natural compounds we collect. Some of the compounds we tested are also still a little literature that discusses them in detail. However, research and development of antiviral compounds derived from natural compounds from animals, plants, and microorganisms are crucial for finding and developing anti-dengue drugs that are more effective and safer to use. Therefore, the opportunities in this field of research are still very wide open because DENV infection is one of the health problems that are a priority of WHO. This research is a preliminary study so that natural compounds which are considered potential in this study must be developed and further tested through in vitro and in vivo research which is then followed by drug toxicity testing and clinical trials in order to meet the eligibility standard as an effective anti-dengue drug before really applied to humans. Further development and research in the future can also help to optimize these compounds as active anti-dengue compounds. In the process of research on potential natural compounds as anti-dengue, exploration of the mechanism of action of potential compounds, especially the halishigamide A against DENV NS5, is needed to support the development of this compound so that it can be expected to solve the problem of dengue infection in the future.
CONCLUSION
Nine compounds are expected to be strong candidates for antiviral compounds that target DENV2 NS5 based on in silico analysis. They are halishigamide A; trigocherriolide A and C; quinadoline A and B; zingiberene; and tryptoquivaline E, F, and H. However, the strongest candidate in this study is halishigamide A. It has an affinity value twice as high as ribavirin. It also has a binding site and bond with the residues that are relatively identical as ribavirin. Also, it is bonded more dynamically than ribavirin. Further studies needed to confirm this study's prediction and further development of these compounds as antiviral compounds for DENV.
REFERENCES
Allard, P.M., Leyssen, P., Martin, M.T., Bourjot, M., Dumontet, V., Eydoux, C., Guillemot, J.C., Canard, B., Poullain, C., Guéritte, F., et al. 2012. Antiviral chlorinated daphnane diterpenoid orthoesters from the bark and wood of Trigonostemon cherrieri. Phytochemistry. 84: 160–168.
Andjelic, C., Planelles, V., and Barrows, L. 2008. Characterizing the anti-HIV activity of papuamide a. Marine Drugs. 6: 528–549.
Apriyanto, D.R., Aoki, C., Hartati, S., Arsianti, A., Louisa, M., and Hotta, H. 2016. Aktivitas antivirus hepatitis c fraksi n-heksana, etil asetat, dan n-butanol daun lengkeng (Dimocarpus longan LOUR). Prosiding Seminar Nasional Hasil-Hasil PPM IPB 2016. 11.
Ashour, J., Laurent-Rolle, M., Shi, P.Y., and García-Sastre, A. 2009. NS5 of dengue virus mediates STAT2 binding and degradation. Journal of Virology. 83: 5408–5418.
Atun, S. 2009. Potensi senyawa isoflavon dan derivatnya dari kedelai (Glycine max. L) serta manfaatnya untuk kesehatan. Prosiding Seminar Nasional Penelitian, Pendidikan dan Penerapan MIPA, Fakultas MIPA, Universitas Negeri Yogyakarta. 9.
Balafif, R.A.R., Andayani, Y., and Gunawan, E.R. 2019. Analisis senyawa triterpenoid dari hasil fraksinasi ekstrak air buah buncis (Phaseolus vulgaris linn). Chemistry progress. 6.
Bhatt, S., Gething, P.W., Brady, O.J., Messina, J.P., Farlow, A.W., Moyes, C.L., Drake, J.M., Brownstein, J.S., Hoen, A.G., Sankoh, O., et al. 2013. The global distribution and burden of dengue. Nature. 496: 504–507.
Bone, K. and Mills, S. 2013. Principles and practice of phytotherapy: Modern herbal medicine 2nd ed. Elsevier: Churchill Livingstone.
Bourjot, M., Leyssen, P., Neyts, J., Dumontet, V., and Litaudon, M. (2014). Trigocherrierin A, a potent inhibitor of chikungunya virus replication. Molecules. 19: 3617–3627.
Bryskier, A. and Bergogne-Bérézin, E. 2005. Macrolides. In Bryskier, M.D., editor. Antimicrobial agents. American Society of Microbiology. p. 475–526.
Chung, H.J. 2009. Evaluation of the biological activity of extracts from Star-Anise (Illicium verum). Preventive Nutrition and Food Science. 14: 195–200.
Dewi, N.A.K., and Subarnas, A. 2018. Efektivitas beberapa jenis tanaman sebagai antivirus flu burung (avian influenza). Farmaka. 16: 1–12.
Diamond, M.S. and Pierson, T.C. 2015. Molecular insight into dengue virus pathogenesis and its implications for disease control. Cell, 162: 488–492.
Donia, M. and Hamann, M.T. 2003. Marine natural products and their potential applications as anti-infective agents. The Lancet Infectious Diseases. 3: 338–348.
Dubankova, A. and Boura, E. 2019. Structure of the yellow fever NS5 protein reveals conserved drug targets shared among flaviviruses. Antiviral Research. 169: 104536.
El Sahili, A. and Lescar, J. 2017. Dengue virus non-structural protein 5. Viruses. 9.
Farhaty, N. and Muchtaridi, M. 2016. Tinjauan kimia dan aspek farmakologi senyawa asam klorogenat pada biji kopi: review. Farmaka. 14: 214–227.
García, L.L., Padilla, L. and Castaño, J.C. 2017. Inhibitors compounds of the flavivirus replication process. Virology Journal. 14.
Guo, Z., Gai, C., Cai, C., Zeng, Y., Yuan, J., Mei, W., and Dai, H. 2018. Quinadoline C, a new alkaloid from mangrove Sonneratia hainanensis -associated Aspergillus sp. Journal of Shenzhen University Science and Engineering. 35: 334.
Hart, D.J. 2010. The spiroquinazoline family of alkaloids: a review. 34.
Haryanto, B. 2018. Indonesia dengue fever: status, vulnerability, and challenges. In Rodriguez-Morales, A.J., editor. Current Topics in Tropical Emerging Diseases and Travel Medicine. IntechOpen.
Herman, R. 2019. Studi in silico lima senyawa aktif sebagai penghambat protein virus dengue. Jurnal Kefarmasian Indonesia. 40–47.
Jayaprakash, V., Castagnolo, D., and Özkay, Y. 2019. Medicinal chemistry of neglected and tropical diseases: advances in the design and synthesis of antimicrobial agents. CRC Press.
Karpiński, T.M. 2019. Marine macrolides with antibacterial and/or antifungal activity. Marine Drugs. 17: 241.
Karyanti, M. R. and Hadinegoro, S. R. 2016. Perubahan epidemiologi demam berdarah dengue di indonesia. Sari Pediatri. 10: 424–432.
Karyawati, A.T. 2011. Aktivitas antivirus simian retrovirus serotype-2 (srv-2) dari ekstrak meniran (Phyllanthus niruri) dan temu lawak (Curcuma Xanthorrhiza). Jurnal Penelitian Sains. 14.
Klema, V.J., Ye, M., Hindupur, A., Teramoto, T., Gottipati, K., Padmanabhan, R., and Choi, K.H. 2016. Dengue virus nonstructural protein 5 (ns5) assembles into a dimer with a unique methyltransferase and polymerase interface. PLoS Pathogens. 12: e1005451.
Koswandy, L.F. and Ramadhania, Z.M. 2016. Kandungan senyawa kimia dan bioaktivitas dari eucalyptus globulus labill. Farmaka. 14: 63–78.
Koyama, N., Inoue, Y., Sekine, M., Hayakawa, Y., Homma, H., O̅mura, S., and Tomoda, H. 2008. Relative and absolute stereochemistry of quinadoline b, an inhibitor of lipid droplet synthesis in macrophages. Organic Letters. 10: 5273–5276.
Lee, J.C., Chang, F.R., Chen, S.R., Wu, Y.H., Hu, H.C., Wu, Y.C., Backlund, A., and Cheng, Y.B. 2016. Anti-dengue virus constituents from Formosan Zoanthid Palythoa mutuki. Marine Drugs. 14: 151.
Leneva, I.A., Russell, R.J., Boriskin, Y.S., and Hay, A.J. 2009. Characteristics of arbidol-resistant mutants of influenza virus: Implications for the mechanism of anti-influenza action of arbidol. Antiviral Research. 81: 132–140.
Lim, S.P., Koh, J.H.K., Seh, C.C., Liew, C.W., Davidson, A.D., Chua, L.S., Chandrasekaran, R., Cornvik, T.C., Shi, P.Y., and Lescar, J. 2013. A crystal structure of the dengue virus non-structural protein 5 (ns5) polymerase delineates interdomain amino acid residues that enhance its thermostability and de novo initiation activities. The Journal of Biological Chemistry. 288:31105–31114.
Lipinski, C.A. 2004. Lead- and drug-like compounds: the rule-of-five revolution. Drug Discovery Today: Technologies. 1: 337–341.
Mao, Q.Q., Xu, X.Y., Cao, S.Y., Gan, R.Y., Corke, H., Beta, T., and Li, H.B. 2019. Bioactive compounds and bioactivities of ginger (Zingiber officinale roscoe). Foods. 8: 185.
Masyeni, S., Yohan, B., and Sasmono, R.T. 2019. Concurrent infections of dengue virus serotypes in Bali, Indonesia. BMC Research Notes. 12: 129.
Matsunaga, S. 2006. Trisoxazole macrolides from Hexabranchus nudibranchs and other marine invertebrates. In Cimino, C., and Gavagnin, M., editors. Molluscs. Progress in Molecular and Subcellular Biology. 43. Berlin: Springer.
Menninger, J.R. 1995. Mechanism of inhibition of protein synthesis by macrolide and lincosamide antibiotics. Journal of Basic and Clinical Physiology and Pharmacology. 6: 229–250.
Mentari, D. and Wijayanti, S. 2016. Aktivitas antivirus ekstrak n-heksan Streptomyces sp. GMR22 terhadap virus dengue serotipe 1.
Moghadamtousi, S., Nikzad, S., Kadir, H., Abubakar, S., and Zandi, K. 2015. Potential antiviral agents from marine fungi: an overview. Marine Drugs. 13: 4520–4538.
Nadjib, M., Setiawan, E., Putri, S., Nealon, J., Beucher, S., Hadinegoro, S.R., Permanasari, V.Y., Sari, K., Wahyono, T.Y.M., Kristin, E., et al. 2019. Economic burden of dengue in Indonesia. PLoS Neglected Tropical Diseases. 13: e0007038.
Nyström, K., Waldenström, J., Tang, K.W., and Lagging, M. 2019. Ribavirin: pharmacology, multiple modes of action and possible future perspectives. Future Virology. 14: 153–160.
Oliveira, A.S. de, Silva, M.L. da, Oliveira, A.F.C.S., Silva, C.C. da, Teixeira, R.R., and De Paula, S.O. 2014. NS3 and NS5 proteins: important targets for anti-dengue drug design. Journal of the Brazilian Chemical Society. 25: 1759–1769.
Potisopon, S., Priet, S., Collet, A., Decroly, E., Canard, B., and Selisko, B. 2014. The methyltransferase domain of dengue virus protein NS5 ensures efficient RNA synthesis initiation and elongation by the polymerase domain. Nucleic Acids Research. 42: 11642–11656.
Rajapakse, S. 2011. Dengue shock. Journal Emerg Trauma Shock. 4: 120.
Ramharack, P. and Soliman, M.E.S. 2018. Zika virus NS5 protein potential inhibitors: An enhanced in silico approach in drug discovery. Journal of Biomolecular Structure and Dynamics. 36: 1118–1133.
Roy, B.G. 2017. Potential of small-molecule fungal metabolites in antiviral chemotherapy. Antiviral Chemistry and Chemotherapy. 25: 20–52.
Sehnal, D., Rose, A., Koca, J., Burley, S., and Velankar, S. 2018. Mol*: towards a common library and tools for web molecular graphics. Workshop on Molecular Graphics and Visual Analysis of Molecular Data. p. 5.
Senthilvel, P., Lavanya, P., Kumar, K.M., Swetha, R., Anitha, P., Bag, S., Sarveswari, S., Vijayakumar, V., Ramaiah, S., and Anbarasu, A. 2013. Flavonoid from Carica papaya inhibits NS2B-NS3 protease and prevents Dengue 2 viral assembly. Bioinformation. 9: 889–895.
Shang, X.F., Morris-Natschke, S.L., Yang, G.Z., Liu, Y.Q., Guo, X., Xu, X.S., Goto, M., Li, J.C., Zhang, J.Y., and Lee, K.H. 2018. Biologically active quinoline and quinazoline alkaloids part II. Medicinal Research Reviews. 38: 1614–1660.
Shimizu, H., Saito, A., Mikuni, J., Nakayama, E.E., Koyama, H., Honma, T., Shirouzu, M., Sekine, S., and Shioda, T. 2019. Discovery of a small molecule inhibitor targeting dengue virus NS5 RNA-dependent RNA polymerase. PLoS Neglected Tropical Diseases. 13: e0007894.
Sulistyawati, S., Dwi Astuti, F., Rahmah Umniyati, S., Tunggul Satoto, T.B., Lazuardi, L., Nilsson, M., Rocklov, J., Andersson, C., and Holmner, Å. 2019. Dengue vector control through community empowerment: lessons learned from a community-based study in Yogyakarta, Indonesia. International Journal of Environmental Research and Public Health. 16.
Tm, K. 2019. Marine macrolides with antibacterial and/or antifungal activity. Marine Drugs. 17.
Trott, O. and Olson, A.J. 2009. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, NA-NA.
Walsham, G. 2020. Health information systems in developing countries: Some reflections on information for action. Information Technology for Development. 26: 194–200.
Warkentien, T. 2016. Dengue fever: historical perspective and the global response. Journal of Infectious Diseases and Epidemiology. 2.
World Health Organization, and Department of Control of Neglected Tropical Diseases. 2017. Integrating neglected tropical diseases into global health and development: Fourth WHO report on neglected tropical diseases. World Health Organization.
Wu, J., Liu, W., and Gong, P. 2015. A structural overview of RNA-dependent RNA polymerases from the flaviviridae family. International Journal of Molecular Sciences. 16: 12943–12957.
Wu, M. and Ma, D. 2013. Total Syntheses of (±)-Spiroquinazoline, (−)-Alantryphenone, (+)-Lapatin A, and (−)-Quinadoline B. Angewandte Chemie International Edition. 52: 9759–9762.
Zahra, S. and Iskandar, Y. 2017. Review Artikel :kandungan senyawa kimia dan bioaktivitas Ocimum Basilicum L. Farmaka. 15: 143–152.
Zhang, H., Tang, Y., Ruan, C., and Bai, X. 2016. Bioactive secondary metabolites from the endophytic Aspergillus genus. Records of Natural Products. 16.
Zhang, T., Wang, M.L., Zhang, G.R., Liu, W., Xiao, X.-Q., Yang, Y.S., Li, J.T., Xun, Z.M., Li, D.Y., and Chan, P.K.S. 2019. Recombinant DENV 2 NS5: an effective antigen for diagnosis of DENV infection. Journal of Virological Methods. 265: 35–41.
OPEN access freely available online
Chiang Mai University Journal of Natural Sciences [ISSN 16851994]
Chiang Mai University, Thailand
https://cmuj.cmu.ac.th
Arief Hidayatullah1, Wira Eka Putra1,2,* , Wa Ode Salma3, Bayyinatul Muchtaromah4, Galuh Wening Permatasari5, Hendra Susanto1,2, Diana Widiastuti6, and Muhammad Kismurtono7
1 Department of Biology, Faculty of Mathematics and Natural Sciences, Universitas Negeri Malang, Malang 65145, Indonesia
2 Department of Biotechnology, Faculty of Mathematics and Natural Sciences, Universitas Negeri Malang, Malang 65145, Indonesia
3 Department of Nutrition, Faculty of Public Health, Halu Oleo University, Indonesia
4 Department of Biology, Faculty of Science and Technology, Universitas Islam Negeri Maulana Malik Ibrahim, Malang 65144, Indonesia
5 Indonesian Research Institute for Biotechnology and Bioindustry, Bogor, Indonesia
6 Department of Chemistry, Faculty of Mathematics and Natural Science, Universitas Pakuan, Indonesia
7 Research Unit for Natural Product Technology, The Indonesian Institute of Science, Indonesia
Corresponding author: Wira Eka Putra, E-mail: wira.putra.fmipa@um.ac.id
Total Article Views
Editor: Wasu Pathom-aree,
Chiang Mai University, Thailand
Article history:
Received: June 24, 2020;
Revised: September 11, 2020;
Accepted: October 22, 2020

