
Unique Genetic Structure of Y-chromosomal Lineage of the Moken from the Andaman Sea of Thailand
Tanapon Seetaraso, Wibhu Kutanan, Jatupol Kampuansai, Kanha Muisuk, and Metawee Srikummool*Published Date : 2020-09-01
DOI : https://doi.org/10.12982/CMUJNS.2020.0066
Journal Issues : Number 4, October-December 2020
ABSTRACT To investigate the genetic structure of the Y-chromosome of the Moken people living in the Andaman Sea area of Thailand, and to compare the genetic relationship between them and other groups on the mainland, as well as the islands Southeast Asia. This study analyzed 17 Y-chromosomal short tandem repeat (Y-STR) loci in 11 unrelated Moken males from Ranong Province of Thailand. The genomic DNA was extracted from buccal cells, and amplified using an AmpFℓSTR Yfiler PCR Amplification Kit. We compared Y-STR genotypes with other populations from the Y Chromosome Haplotype Reference Database (YHRD) and constructed the multidimensional scaling (MDS) plot and the neighbor-joining tree (NJ), based on genetic distance Rst. Nine different haplotypes from 11 unrelated individuals were identified. The haplotype diversity was 0.9455 ± 0.066, which was closer to other Southeast Asian populations. The genetic relationship results also showed the genetic differentiation of the Moken, without any significant differences among them, as well as some Austronesian (AN) groups from Malaysia and Sino-Tibetan (ST) groups from Taiwan. This study suggests that there was a unique genetic structure on the Y-chromosome of the Moken from the Andaman Sea, showing genetic relations with some AN and ST groups.
Keywords: Y chromosomal-STR, Genetic structure, Moken, Andaman Sea
INTRODUCTION
The Moken is one of the sea nomad groups, who inhabit the coastal area and islands of the Andaman Sea, in the southern part of Thailand, and are distributed up the coast of the Mergui Archipelago of Myanmar. The ancestors of the Moken people might have lived on the Southeast Asian (MSEA) mainland which is now Myanmar, and spread southwards to Malaysia (Hinshiranan, 1996). The alternative in hypothesis of the origin of the Moken is that they originated in the islands of Southeast Asia (ISEA) (Koechlin, 1967). However, genetic evidence, supports the hypothesis that the Moken originated within the coastal region of MSEA before migrating to ISEA (Dancause et al., 2009). Thus, the origins of the Moken are still debated.
Mitochondrial DNA (mtDNA) and Y-chromosomal DNA are two uniparental genetic markers, which are commonly used to reconstruct population history, and investigate the genetic relationship among populations, including the tracing of lineages of populations (Richards et al., 2000; Maca-Meyer et al., 2001; Y Chromosome Consortium, 2002; Jobling and Tyler, 2003; Barbieri et al., 2013). mtDNA is maternally inherited, while the Y-chromosome is a useful tool to study paternal ancestry, because it is passed down from father to son and the majority of its DNA lacks recombination (Y Chromosome ; Jobling and Tyler, 2003; Santana et al., 2014). A previous study of the mtDNA variation of the Moken living in Phuket province of Thailand indicated a low mitochondrial diversity and distract genetic structure from Austronesian speaking groups and the MSEA people (Lum et al., 1998). Another study of mtDNA variation of the Moken, who live in the Mergui Archipelago of Myanmar, reported the basal mtDNA lineage of the Moken; observed M21b2 and M46a (Dancause et al., 2009). A recent mtDNA study of the Moken from Chang Island suggested the signal of the influence of genetic drift. The identified mitochondrial lineages were M21b2, D4e1a, M46a and F1a1c1. The genetic relationship analysis suggested that the Moken on Chang Island shared common ancestors with the Moken people in the Mergui Archipelago and with some Austroasiatic speaking groups in MSEA. It also suggested that the Moken originated on the MSEA before migration to the islands (Seetaraso et al., 2019). However, the study of paternal lineage of the Moken people have not been studied.
In this study, we aim to investigate the genetic structure of Y-chromosomal DNA in the Moken people from Andaman Sea of Thailand and compare the genetic relationship between them and other published data. We examined the genotype 17 Y-chromosomal STR loci using AmpFℓSTR Yfiler PCR Amplification Kit (Applied Biosystems, USA), from 11 Moken males. We found a unique genetic structure of Y-chromosomal lineage in the male Moken.
MATERIAL AND METHODS
Studied populations
This study was approved by the Naresuan University Institutional Review Board (COA No. 0464/2017). Buccal samples were collected with informed consent from 11 unrelated male volunteers from Chang Island in the Ranong Province of Thailand (TH-MG) (Figure 1). The recruitment criteria for volunteers was unrelated men belonging to the Moken from Chang Island having at least two generations of ancestors, without any genetic admixed by other tribes. DNA was extracted from the buccal cells with the Gentra Puregene Buccal Cell Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocols. Sample DNA quantity and purity were determined by NanoDrop 2,000 UV-Vis Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The Y-chromosomal STR data from 59 populations on the mainland as well as the islands of Southeast Asia (Table 1), were taken from the Y-chromosomal STR Haplotype Reference Database (YHRD; http://www.yhrd.org/) () to then be compared with our results.
Figure 1. Map of sampling location of the Moken in Chang Island, Ranong Province of Thailand.
Table 1. General information about the current studied and other populations from the YHRD database.
|
No. |
Country |
Population |
Code |
Linguistic family |
Size (n) |
Accession number |
No. |
Country |
Population |
Code |
Linguistic family |
Size (n) |
Accession number |
|
1 |
Thailand |
Moken |
TH-MG |
AN |
11 |
This study |
31 |
Malaysia |
MY-BY |
Bidayuh |
AN |
113 |
YA003415 |
|
2 |
Thailand |
Thai |
TH-TH |
TK |
140 |
YA004250 |
32 |
Malaysia |
MY-CH |
Champa |
AN |
13 |
YA004033 |
|
3 |
Thailand |
Central Thai |
TH-CE |
TK |
256 |
YA003923 |
33 |
Malaysia |
MY-IB |
Iban |
AN |
103 |
YA003416 |
|
4 |
Thailand |
Northern Thai |
TH-NT |
TK |
15 |
YA004164 |
34 |
Malaysia |
MY-JA |
Jawa |
AN |
14 |
YA004039 |
|
5 |
Thailand |
Padong |
TH-PD |
ST |
13 |
YA004162 |
35 |
Malaysia |
MY-KD |
Kedah |
AN |
9 |
YA004035 |
|
6 |
Thailand |
Pwo |
TH-PW |
ST |
15 |
YA004168 |
36 |
Malaysia |
MY-KL |
Kelantan |
AN |
43 |
YA004038 |
|
7 |
Thailand |
Shan |
TH-SH |
TK |
12 |
YA004163 |
37 |
Malaysia |
MY-KS |
Kensiu |
AA |
16 |
YA004133 |
|
8 |
Thailand |
Skaw |
TH-SK |
ST |
24 |
YA004167 |
38 |
Malaysia |
MY-LA |
Lanoh |
AA |
11 |
YA004134 |
|
9 |
Thailand |
Yong |
TH-YO |
TK |
124 |
YA004161 |
39 |
Malaysia |
MY-ME |
Melanau |
AN |
104 |
YA003417 |
|
10 |
Bangladesh |
BD-MA |
Barma |
ST |
107 |
YA004285 |
40 |
Malaysia |
Minang |
MY-MI |
AN |
23 |
YA004036 |
|
11 |
Bangladesh |
BD-RA |
Rakhine |
ST |
110 |
YA004284 |
41 |
Malaysia |
Northern Borneo |
MY-NB |
AN |
51 |
YA003927 |
|
12 |
China |
CN-HA |
Han |
ST |
30 |
YA003866 |
42 |
Malaysia |
Rawa |
MY-RA |
AN |
11 |
YA004034 |
|
13 |
China |
CN-FJH |
Han |
ST |
124 |
YA004397 |
43 |
Malaysia |
Semai |
MY-SE |
AA |
18 |
YA004135 |
|
14 |
China |
CN-FJS |
She |
ST |
92 |
YA004031 |
44 |
Philippines |
Filipino |
PH-FP |
AN |
943 |
YA002895 YA003206 YA003892 |
|
15 |
China |
CN-FZD |
Danmin |
ST |
87 |
YA004488 |
45 |
Philippines |
Manila |
PH-MA |
AN |
64 |
YA003202 |
|
16 |
China |
CN-FZS |
She |
ST |
152 |
YA003747 YA004066 |
46 |
Singapore |
Malay |
SG-ML |
AN |
556 |
YA004367 YA003272 |
|
17 |
China |
CN-GDH |
Han |
ST |
1623 |
YA004066 YA004304 YA004330 YA004392 |
47 |
Taiwan |
Ami |
TW-AM |
AN |
86 |
YA003920 YA003961 |
|
18 |
China |
CN-GXG |
Gin |
AA |
161 |
YA004217 |
48 |
Taiwan |
Atayal |
TW-AT |
AN |
240 |
YA003524 YA003919 YA003964 |
|
19 |
China |
CN-GXM |
Miao |
AA |
186 |
YA004220 |
49 |
Taiwan |
Bunan |
TW-BU |
AN |
38 |
YA003918 |
|
20 |
China |
CN-GXY |
Yao |
AA |
138 |
YA004221 |
50 |
Taiwan |
Han |
TW-HA |
ST |
30 |
YA003963 |
|
21 |
China |
CN-GXZ |
Zhuang |
TK |
2913 |
YA003542 YA004006 YA004208 YA004222 |
51 |
Taiwan |
Hakka |
TW-HK |
ST |
22 |
YA003962 |
|
22 |
China |
CN-SZH |
Han |
ST |
136 |
YA004280 |
52 |
Taiwan |
Paiwan |
TW-PA |
AN |
237 |
YA003500 YA003915 |
|
23 |
China |
CN-XBD |
Dai |
TK |
92 |
YA003876 YA003877 |
53 |
Taiwan |
Puyama |
TW-PU |
AN |
15 |
YA003913 |
|
24 |
Indonesia |
IN-BT |
Batak |
AN |
100 |
YA003769 |
54 |
Taiwan |
Rukai |
TW-RU |
AN |
28 |
YA003916 |
|
25 |
Indonesia |
IN-JA |
Java |
AN |
137 |
YA003200 YA003770
|
55 |
Taiwan |
Saisiat |
TW-SA |
AN |
24 |
YA003914 |
|
26 |
Lao |
LA-LT |
Laotian |
TK |
45 |
YA003948 |
56 |
Taiwan |
Tsou |
TW-TS |
AN |
27 |
YA003912 |
|
27 |
Malaysia |
MY-AC |
Achen |
AN |
7 |
YA004032 |
57 |
Taiwan |
Yami |
TW-YA |
AN |
24 |
YA003911 |
|
28 |
Malaysia |
MY-BJ |
Banjar |
AN |
18 |
YA004040 |
58 |
Vietnam |
Kinh |
VN-KI |
AA |
123 |
YA003984 |
|
29 |
Malaysia |
MY-BT |
Bateq |
AA |
19 |
YA004131 |
59 |
Vietnam |
Mong |
VN-MO |
AA |
59 |
YA003985 |
|
30 |
Malaysia |
MY-BU |
Bugis |
AN |
15 |
YA004037 |
60 |
Vietnam |
Tay |
VN-TA |
AA |
21 |
YA003986 |
Note: BD: Bangladesh, CN: China, IN: Indonesia, MY: Malaysia, PH: Philippines, SG: Singapore, TH: Thailand, TW: Taiwan, VN: Vietnam.
Y-STR genotyping
All samples were genotyped for 17 Y-chromosomal STR loci (DYS19, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, DYS385a/b, DYS438, DYS439, DYS437, DYS448, DYS456, DYS458, DYS635, and Y-GATA H4) with the AmpFℓSTR Yfiler PCR Amplification Kit, according to the manufacturer’s recommendations. Amplification reactions were performed using a GeneAmp PCR System 9700: these products were analyzed in an ABI 3130 sequencer (Applied Biosystems). Genotyping designations were based on a comparison with allelic ladders in the AmpFℓSTR Yfiler PCR Amplification Kit, along with GeneMapper ID-X software (Applied Biosystems).
Statistical analyses
The allele and haplotype frequencies were counted directly, and then compared. Each of the haplotypes were found with the YHRD database. Gene diversity (GD) of the 17 Y-chromosomal STR loci and haplotype diversity (h) (Nei, 1987), were calculated using the ARLEQUIN software package v. 3.5.2.2 (Bern, Switzerland) (Excoffier and Lischer, 2010). We also compared haplotype diversity between the Moken and other MSEA and ISEA populations, according to four language families: Austronesian (AN), Austroasiatic (AA), Tai-Kadai (TK), and Sino-Tibetan (ST) from previous studies (Trejaut et al., 2014; Brunelli et al., 2017; RahayuKarmilla et al., 2018).
To ascertain the genetic relationship among populations, we compared the 17 Y-chromosomal STR loci with the published data of 59 Southeast Asian populations, belonging to four linguistic categories (AN, AA, TK, and ST) in the YHRD database (Table 1). The genetic distance (Rst) between pairs of the population and their statistical significance using 10,000 permutations, were estimated by AMOVA and MDS tools of the YHRD. The genetic distance was plotted in two dimensions by means of a multidimensional scaling (MDS) plot, using the XLSTAT statistical software demo version (Addinsoft, Paris, France). A neighbor-joining (NJ) tree was constructed using the MEGA-X (Kumar et al., 2018).
RESULTS
Allele frequencies, gene diversities, and haplotype diversity
Given 11 unrelated Moken men, they were analyzed for allele frequency distributions and gene diversities of the 17 Y-chromosomal STR loci (Table 2). A total of 54 alleles from 17 Y-chromosomal STR loci were identified with the corresponding allele frequencies, ranking from 0.0909 to 0.9091. The DYS448 is the highest polymorphic STR locus (7 alleles), and showed the highest gene diversity (GD = 0.9091), whereas the lowest polymorphic STR locus was DYS391 (2 alleles and GD = 0.1818).
Among 11 males, there are nine haplotypes (Table 3), out of which eight were single unique, and one (HAP03) was multiple unique. The haplotype diversity was found to be 0.9455 ± 0.066. When we compared the haplotypes of the Moken with other people from the YHRD database, we found that one haplotype (HAP08) exactly matched that of the ST people from China. The haplotype diversity of the Moken was close to the values from various comparative populations (Trejaut et al, 2014; Brunelli et al, 2017; RahayuKarmilla et al; 2018) (Figure 2).
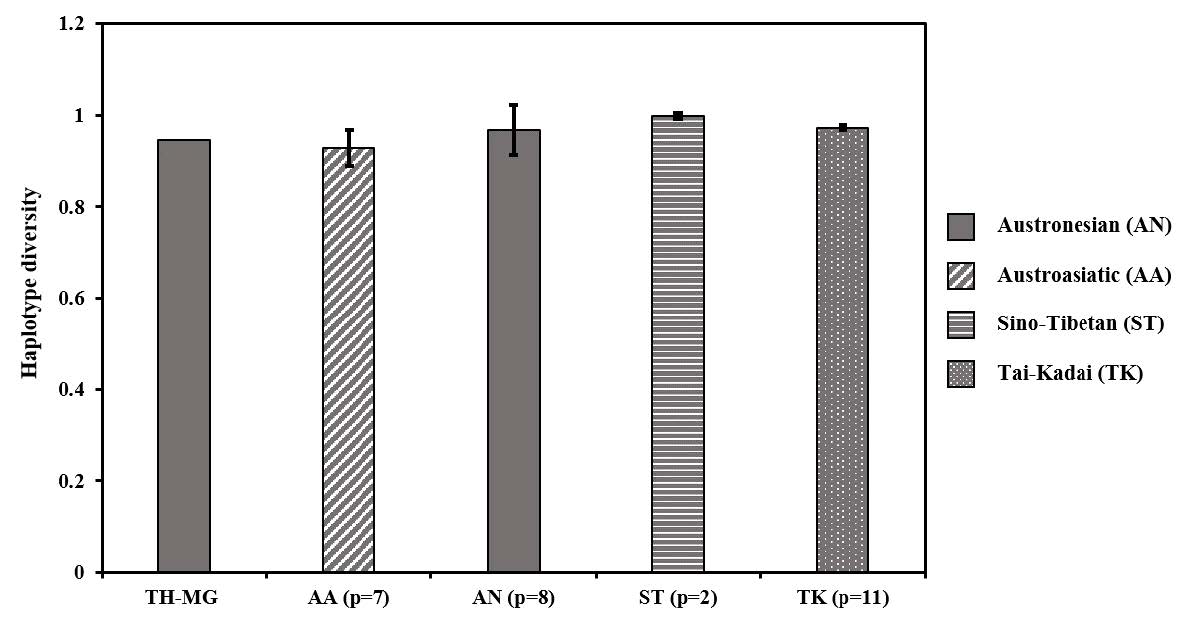
Figure 2. Comparison of haplotype diversity between the Moken and other Southeast Asian people. The error bars show range of haplotype diversity within linguistic family. (p = the number of populations).
Table 2. Allele frequencies and gene diversities of the 17 Y-chromosomal STR loci of the Moken
|
Allele |
DYS19 |
DYS389l |
DYS389ll |
DYS390 |
DYS391 |
DYS392 |
DYS393 |
DYS437 |
DYS438 |
DYS439 |
DYS448 |
DYS456 |
DYS458 |
DYS635 |
YGATA H4 |
Genotype |
DYS385 |
|
9 |
|
|
|
|
|
|
|
|
0.4545 |
|
|
|
|
|
|
11, 16 |
0.1111 |
|
10 |
|
|
|
|
0.9091 |
|
|
|
0.2727 |
|
|
|
|
|
|
12, 12 |
0.1111 |
|
11 |
|
0.0909 |
|
|
0.0909 |
0.0909 |
|
|
0.2727 |
0.4545 |
|
|
|
|
0.4545 |
13, 15 |
0.2222 |
|
12 |
|
0.1818 |
|
|
|
0.0909 |
0.3636 |
|
|
0.2727 |
|
|
|
|
0.4545 |
13, 20 |
0.1111 |
|
13 |
|
0.2727 |
|
|
|
0.4545 |
0.6364 |
|
|
0.2727 |
|
|
|
|
0.0909 |
13, 21 |
0.2222 |
|
14 |
0.7273 |
0.4545 |
|
|
|
0.3636 |
|
0.3636 |
|
|
|
|
|
|
|
14, 19 |
0.2222 |
|
15 |
0.2727 |
|
|
|
|
|
|
0.4545 |
|
|
0.2727 |
0.7273 |
|
|
|
|
|
|
16 |
|
|
|
|
|
|
|
0.1818 |
|
|
0.1818 |
0.0909 |
0.2727 |
|
|
|
|
|
17 |
|
|
|
|
|
|
|
|
|
|
|
0.1818 |
0.2727 |
|
|
|
|
|
18 |
|
|
|
|
|
|
|
|
|
|
0.1818 |
|
0.1818 |
|
|
|
|
|
19 |
|
|
|
|
|
|
|
|
|
|
0.0909 |
|
|
0.2727 |
|
|
|
|
20 |
|
|
|
|
|
|
|
|
|
|
0.0909 |
|
0.1818 |
0.4545 |
|
|
|
|
21 |
|
|
|
0.2727 |
|
|
|
|
|
|
0.0909 |
|
0.0909 |
0.1818 |
|
|
|
|
22 |
|
|
|
0.4545 |
|
|
|
|
|
|
0.0909 |
|
|
|
|
|
|
|
23 |
|
|
|
0.2727 |
|
|
|
|
|
|
|
|
|
0.0909 |
|
|
|
|
27 |
|
|
0.0909 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
28 |
|
|
0.1818 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
29 |
|
|
0.0909 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
30 |
|
|
0.4545 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
32 |
|
|
0.1818 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
GD |
0.4364 |
0.7454 |
0.7818 |
0.7091 |
0.1818 |
0.7091 |
0.5091 |
0.6910 |
0.7091 |
0.7091 |
0.9091 |
0.4727 |
0.8546 |
0.7636 |
0.6367 |
- |
0.9020 |
Note: GD: gene diversity
Table 3. Y-chromosome haplotype distribution of the Moken, based on the 17 Y-chromosomal STR loci.
|
Haplotype |
DYS19 |
DYS385a |
DYS385b |
DYS389l |
DYS398ll |
DYS390 |
DYS391 |
DYS392 |
DYS393 |
DYS437 |
DYS438 |
DYS439 |
DYS448 |
DYS456 |
DYS458 |
DYS635 |
GATA-4 |
Frequency |
|
HAP01 |
15 |
11 |
16 |
12 |
28 |
21 |
10 |
11 |
12 |
14 |
10 |
11 |
21 |
15 |
18 |
23 |
13 |
0.0909 |
|
HAP02 |
15 |
13 |
20 |
11 |
27 |
23 |
10 |
13 |
12 |
15 |
10 |
12 |
22 |
16 |
17 |
21 |
12 |
0.0909 |
|
HAP03 |
14 |
14 |
19 |
14 |
30 |
22 |
10 |
14 |
13 |
14 |
9 |
11 |
15 |
15 |
16 |
19 |
11 |
0.2727 |
|
HAP04 |
14 |
13 |
21 |
13 |
30 |
22 |
11 |
13 |
13 |
15 |
9 |
12 |
16 |
15 |
17 |
20 |
11 |
0.0909 |
|
HAP05 |
14 |
13 |
21 |
13 |
30 |
22 |
10 |
13 |
13 |
15 |
9 |
12 |
16 |
15 |
17 |
21 |
11 |
0.0909 |
|
HAP06 |
15 |
12 |
12 |
13 |
29 |
23 |
10 |
12 |
12 |
15 |
10 |
11 |
19 |
15 |
20 |
20 |
12 |
0.0909 |
|
HAP07 |
14 |
13 |
15 |
14 |
32 |
21 |
10 |
13 |
13 |
16 |
11 |
13 |
18 |
17 |
20 |
20 |
12 |
0.0909 |
|
HAP08 |
14 |
14 |
19 |
12 |
28 |
23 |
10 |
14 |
12 |
15 |
11 |
13 |
20 |
15 |
18 |
20 |
12 |
0.0909 |
|
HAP09 |
14 |
13 |
15 |
14 |
32 |
21 |
10 |
13 |
13 |
16 |
11 |
13 |
18 |
17 |
21 |
20 |
12 |
0.0909 |
Genetic relationship among populations
The genetic distance (Rst) among the population indicated that the Moken population (TH-MG) was not significantly different from the Acheh (MY-AC), the Champa (MY-CH), and the Minang (MY-MI) people from Malaysia, who speak the AN language, along with the ST-speaking Hakka (TW-HK) from Taiwan.
The multidimensional scaling (MDS) plot revealed that the Moken (TH-MG) were separated from other populations, reflecting some degree of differentiation and unique genetic structure. The TK- and ST-speaking groups were clustered together in the center of the plot, indicating their close genetic relationship. The AA- and AN-speaking groups were scattered around the plot indicating genetic heterogeneity. However, eight aboriginal Taiwanese (TW-AM, TW-AT, TW-PA, TW-PU, TW-RK, TW-SA, TW-TS, and TW-YA) were dispersed in the lower right quadrant of the plot, reflecting their divergence (Figure 3).
The NJ tree showed five clusters of population: the Moken population (TH-MG) was not clustered with their linguistic relatives (clusters 3, 4, and 5), but were rather close to the ST group cluster (cluster 1). TK groups were located in cluster 2, while the AA-speaking groups were scattered throughout the tree (Figure 4).
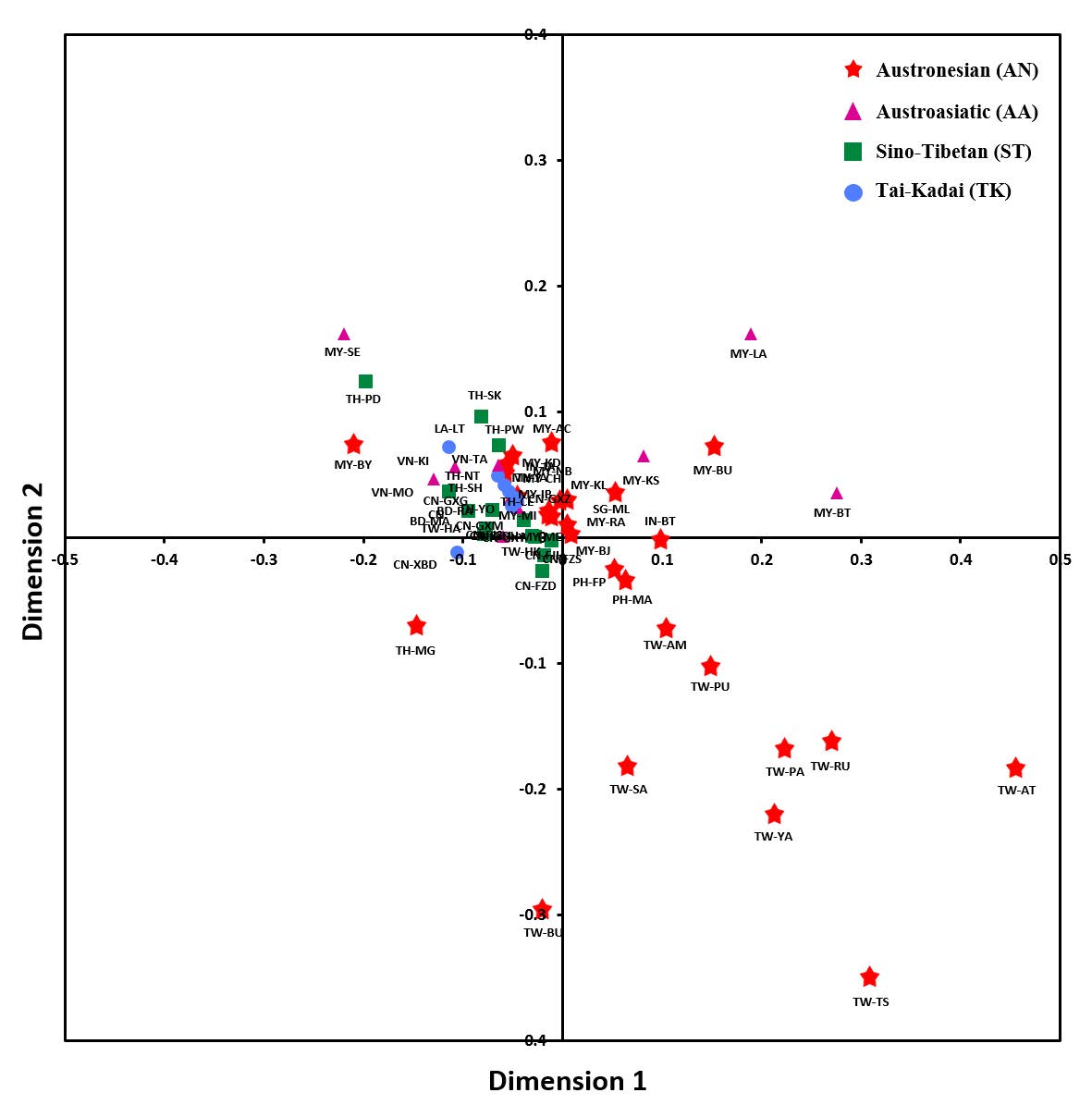
Figure 3. Multidimensional scaling (MDS) plot based on Rst values for 60 populations. Red star, purple triangle, green square and blue circle symbols indicate Austronesian, Austroasiatic, Sino-Tibetan and Tai-Kadai populations, respectively (See abbreviations of populations in Table 1). The stress value is 0.188.
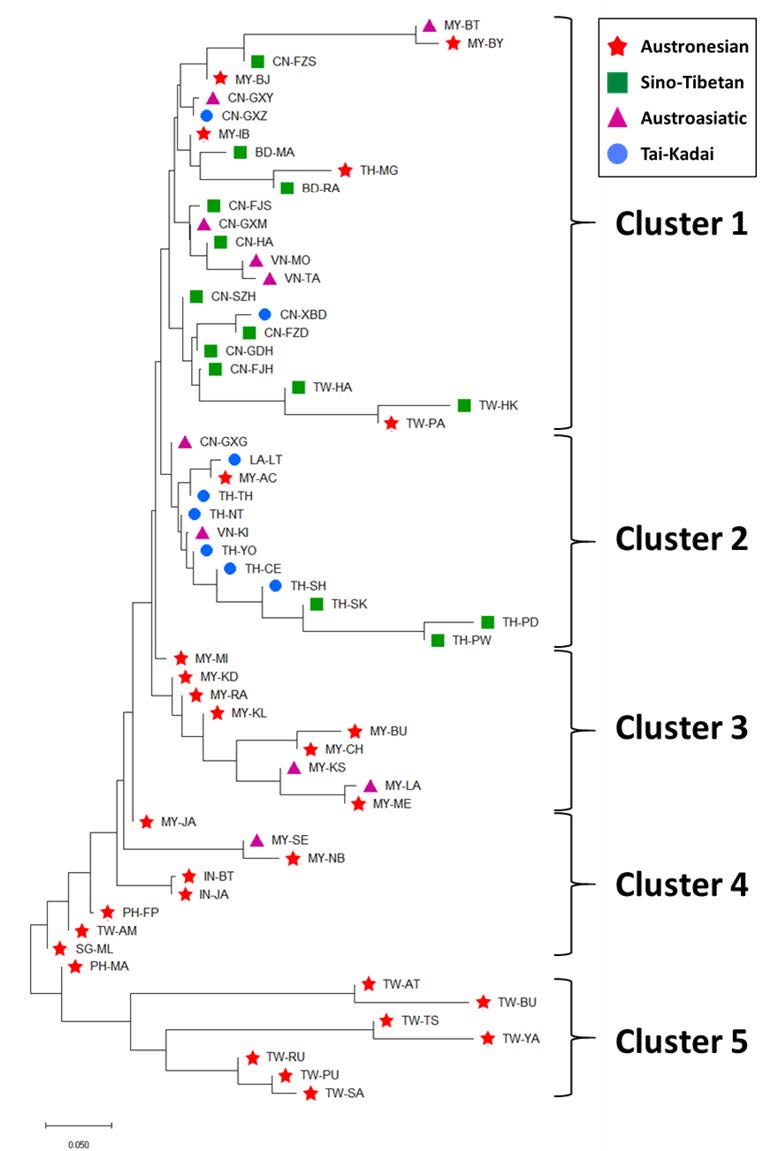
Figure 4. Neighbor-joining (NJ) tree constructed from the Rst values among 60 populations. Red star, purple triangle, green square and blue circle symbols indicate Austronesian, Austroasiatic, Sino-Tibetan and Tai-Kadai population, respectively (See abbreviations of populations in Table 1).
DISCUSSION
This study investigated the genetic structure of 17 Y-chromosome STR loci in 11 Moken males living on Chang Island, originally from Ranong Province, Thailand. The Moken are a group of sea nomads living in the Andaman Sea area who speak a local language, which is classified as an AN-language family. The results show that there are nine haplotypes, with high haplotype diversity (0.9455 ± 0.066), which is close to other Southeast Asian people from previous studies (Trejaut et al., 2014; Brunelli et al., 2017; RahayuKarmilla et al., 2018). The AmpFℓSTR Yfiler PCR Amplification Kit have been used to analyze the Y-chromosome in various populations (Grskovic et al., 2010; Santana et al., 2014). When the identified haplotypes in the Moken were compared with those available from the YHRD database, one was found to match a sample from the Han in China who speak ST languages, thus suggesting that the male Moken may have a genetic relationship with them. This is consistent with other reports about ancestors of the present-day AN population or “Proto-Austronesian speakers,” originating in the area that is now Southeast China (Diamond and Bellwood, 2003; Hwa et al., 2010; Wu et al., 2013).
The genetic relationships between Moken males and those from 59 other Southeast Asian groups showed that the Moken were distantly related to other Austronesian groups, but they tend to relate to certain AN groups in Malaysia and ST groups in Taiwan (Figure 3). The NJ tree suggested that Moken males are distinct from other Austronesian groups; however, they are in a linguistically-mixed cluster, consisting mainly of the ST population (Figure 4). These results show that the Moken male exhibited genetic relationships with both the AN and ST groups, while at the same time, Moken males possess their own unique genetic structure.
A comparison of the Y-chromosomal STR data analyses to the control region in mtDNA data analyses of the same group of Moken (Seetaraso et al., 2019), revealed the difference between their paternal and maternal lineage. Their mitochondrial lineage is close to not only AN, but also to the AA people. As a contradiction, the Y-chromosomal lineage of the Moken is closely related to other AN groups, including a few to ST groups. From this point of view, there are contrasting genetic structures and patterns of female and male lineage in the Moken from Chang Island, Ranong Province. This might result from different microevolutionary forces in male and female lineages. Sex-biased demographic histories of males and females have been reported in previous studies of the Southeast Asian island populations (Kayser et al., 2008; Tumonggor et al., 2013), reflecting an unequal and complex population.
This is one of the studies focusing on the genetic structure of Y-chromosome STR in the Moken of Ranong Province from Thailand. The Moken show a paternal genetic pattern that is unique, but closer to the Austronesian groups of Malaysia and the ST group of Taiwan. These may share a common ancestry, or they may have mixed with them. This study is limited, in that no information on the Y-chromosomal single nucleotide polymorphism (Y-SNP) is included. Further investigation of SNPs will be necessary to identify the origin of the Moken and also that of other Sea Nomadic groups, i.e., the Urak Lawoi and Moken.
ACKNOWLEDGEMENTS
We would like to thank all volunteers and participants who donated their biological samples. This study was funded by Naresuan University (Grant No. R2561B029 and R2562B085). JK acknowledged partially supports by Chiang Mai University, Thailand.
REFERENCES
Barbieri, C., Vicente, M., Rocha, J., Mpoloka, S.W., Stoneking, M., and Pakendorf, B. 2013. Ancient substructure in early mtDNA lineages of southern Africa. The American Journal of Human Genetics. 92(2): 285-292. https://doi.org/10.1016/j.ajhg.2012.12.010
Brunelli, A., Kampuansai, J., Seielstad, M., Lomthaisong, K., Kangwanpong, D., Ghirotto, S., and Kutanan, W. 2017. Y chromosomal evidence on the origin of northern thai people. PLoS One. 12(7): e0181935. https://doi.org/10.1371/journal.pone.0181935
Dancause, K.N., Chan, C.W., Arunotai, N.H., and Lum, J.K. 2009. Origins of the moken sea gypsies inferred from mitochondrial hypervariable region and whole genome sequences. Journal of Human Genetics. 54(2): 86-93.
Diamond, J., and Bellwood, P. 2003. Farmers and their languages: the first expansions. Science. 300(5619): 597-603. https://doi.org/10.1126/science.1078208
Excoffier, L., and Lischer, H.E. 2010. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources. 10(3): 564-567. https://doi.org/10.1111/j.1755-0998.2010.02847.x
Grskovic, B., Mrsic, G., Vrdoljak, A., Merkas, S., Andelinovic, S. 2015. Population genetic analysis of haplotypes based on 17 short tandem repeat loci on Y chromosome in population sample from Eastern Croatia. Croatian Medical Journal. 51(3): 202-208. https://doi.org/10.3325/cmj.2010.51.202
Hinshiranan, N. 1996. The analysis of Moken opportunistic foragers' intragroup and intergroup relations [dissertation]. United States: University of Hawaii.
Hwa, H.L., Tseng, L.H., Ko, T.M., Chang, Y.Y., Yin, H.Y., Su, Y.N., and Lee, J.C. 2010. Seventeen Y-chromosomal short tandem repeat haplotypes in seven groups of population living in Taiwan. Journal of Legal Medicine. 124(4): 295-300. https://doi.org/10.1007/s00414-010-0425-9
Jobling, M.A., and Tyler-Smith, C. 2003. The human Y chromosome: An evolutionary marker comes of age. Nature Reviews Genetics. 4(8):598-612. https://doi.org/10.1038/nrg1124
Kayser, M., Choi, Y.Y., Oven, M.V., Mona, S., Brauer, S., Trent, R.J., Suarkia, D.L., Lehmann, C., and Stoneking, M. 2008. The impact of the Austronesian expansion: evidence from mtDNA and Y chromosome diversity in the admiralty Islands of Melanesia. Molecular biology and evolution. 25(7): 1362-1374. https://doi.org/10.1093/molbev/msn078
Koechlin, B. 1967. The Sea Nomads. A Study Based on the Literature of the Maritime Boat People of Southeast Asia. Lim Bian Han, Singapore.
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution. 35(6): 1547-1549. https://doi.org/10.1093/molbev/msy096
Lum, J.K., Cann, R.L., Martinson, J.J., and Jorde, L.B. 1998. Mitochondrial and nuclear genetic relationships among pacific island and Asian populations. The American Journal of Human Genetics. 63(2): 613-624. https://doi.org/10.1086/301949
Maca-Meyer, N., Gonzalez, A.M., Larruga, J.M., Flores, C., and Cabrera, V.M. 2001. Major genomic mitochondrial lineages delineate early human expansions. BMC genetics. 2: 13. https://doi.org/10.1186/1471-2156-2-13
Nei, M. 1987. Molecular evolutionary genetics. Columbia University Press, New York.
RahayuKarmilla, S.S., Aedrianee, A.R., Haslindawaty, A.R., Azeelah, A.N., Panneerchelvam, S., Norazmi, M.N., and Zafarina, Z. 2018. Paternal lineage affinity of the Malay subethnic and Orang Asli populations in Peninsular Malaysia. International Journal of Legal Medicine. 132(4): 1087-1090. https://doi.org/10.1007/s00414-017-1697-0
Richards, M., Macaulay, V., Hickey, E., Vega, E., Sykes, B., Guida, V., Rengo, C., Sellitto, D., Cruciani, F., Kivisild, T., et al. 2000. Tracing European founder lineages in the near eastern mtdna pool. The American Journal of Human Genetics. 67(5): 1251-1276.
Santana, C., Noris, G., Meraz-Ríos, M.A., Magaña, J.J., Calderon-Aranda, E.S., de Lourdes Muñoz, M., and Gómez, R. 2014. Genetic analysis of 17 Y-STRs in a Mestizo population from the Central Valley of Mexico. Human Biology. 86(4): 289-312. https://doi.org/10.13110/humanbiology.86.4.0289
Seetaraso, T., Kutanan, W., and Srikummool, M. 2019. Mitochondrial DNA variations of the Moken in Ranong Province, Thailand. Proceedings of the 20th National Graduate Research Conference. 2019 Mar 15; Thailand. Khon Kaen, p.469-480.
Trejaut, J.A., Poloni, E.S., Yen, J.C., Lai, Y.H., Loo, J.H., Lee, C.L., He, C.L., and Lin, M. 2014. Taiwan Y-chromosomal DNA variation and its relationship with Island Southeast Asia. BMC Genetics. 15(1):77. https://doi.org/10.1186/1471-2156-15-77
Tumonggor, M.K., Karafet, T.M., Hallmark, B., Lansing, J.S., Sudoyo, H., Hammer, M.F. and Cox, M.P. 2013. The Indonesian archipelago: an ancient genetic highway linking Asia and the Pacific. Journal of Human Genetics. 58(3): 165-173.
Wu, F.C., Chen, M.Y., Chao, C.H., and Pu, C.E. 2013. Study on the genetic polymorphisms of Y chromosomal DNA short tandem repeat loci applied to analyzing the relative affinities among ethnic groups in Taiwan. Forensic Science International: Genetics Supplement Series. 4(1): e69-70. https://doi.org/10.1016/j.fsigss.2013.10.035
Y Chromosome Consortium. 2002. A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Research. 12(2): 339-348. https://doi.org/10.1101/gr.217602
Tanapon Seetaraso1, Wibhu Kutanan2, Jatupol Kampuansai3,4, Kanha Muisuk5, and Metawee Srikummool1*
1Faculty of Medical Science, Naresuan University, Phitsanulok 65000, Thailand
2Faculty of Science, Khon Kaen University, Khon Kaen 40002, Thailand
3Faculty of Science, Chiang Mai University, Chiang Mai 50200, Thailand
4Center of Excellence in Bioresources for Agriculture, Industry and Medicine, Chiang Mai University, Chiang Mai 50200, Thailand
5Faculty of Medicine, Khon Kaen University, Khon Kaen 40002, Thailand
*Corresponding author: metaweesr@nu.ac.th
Total Article Views
Article history:
Received: January 12, 2020
Revised: May 1, 2020
Accepted: May 8, 2020

