
Soluble Expression and Purification of Bioactive Recombinant Human Bone Morphogenetic Protein-2 from Escherichia coli
Waraporn Kasekarn*, Benjawan Suksiriphattanapong, Tawan Chokepaichitkool, Orawan Wanachewin, Sittiruk Roytrakul and Prachya KongtawelertPublished Date : 2020-09-01
DOI : https//:doi.org/10.12982/CMUJNS.2020.0048
Journal Issues : Number 4, October-December 2020
Abstract Human bone morphogenetic protein-2 (hBMP-2) is a potent growth and differentiation factor for bone induction and regeneration. Recombinant hBMP-2 (rhBMP-2) was cloned and expressed as a soluble protein using E. coli-based expression system. A full-length gene encoding mature hBMP-2 was amplified by RT-PCR, cloned into an expression vector and expressed using SHuffle E. coli cells. The rhBMP-2 was successfully expressed as a soluble protein under the control of the lacUV5 and protein A promoters by IPTG induction. The rhBMP-2 fused with ZZ domain at its N-terminus was successively purified with a single step by using IgG Sepharose 6 fast flow affinity chromatography. Analysis of the purified protein on SDS-PAGE, Western blot analysis and LC-MS/MS, verified that the purified protein was rhBMP-2. The biological activity testing on hFOB 1.19 showed that rhBMP-2 had the ability to significantly induce cell proliferation in a dose dependent manner. ALP staining and activity assay also increased after rhBMP-2 treatment. The mRNA expression of the osteogenic genes by quantitative real-time PCR (qRT-PCR) showed that rhBMP-2 was able to up-regulate the gene expression of ALP, COL1, BMP-2, Runx2, and OPN. This data indicates that rhBMP-2 is functionally active to induce human osteoblast proliferation and differentiation. The production of rhBMP-2 by this developed method could be useful for bone regeneration and repair applications.
Keywords: Human bone morphogenetic protein-2, Recombinant protein, Soluble protein, Osteoblast differentiation, Escherichia coli
INTRODUCTION
Bone morphogenetic proteins (BMPs) are the secreted growth factors which belong to the transforming growth factor-b (TGF-b) superfamily of multifunctional cytokines. BMPs have been involved in the regulation of cell proliferation, survival, differentiation, regulation of cell-matrix interactions and the stem cell development in a wide variety of tissues including bone (Hogan, 1996; Lind et al., 1996; Massague, 1996; Nissinen et al., 1997; Balemans and Van Hul, 2002; Xiao et al., 2007; Du and Yip, 2010). BMPs are key regulatory factors in the growth and regeneration of bone and cartilage (Tuan et al., 2003; Ishikawa et al., 2007; Poon et al., 2016), and also function in the repair and remodeling of the adult skeletal system (Sellers et al., 2000). Of the BMP family, BMP-2 is the best characterized protein which has the strongest bone-inducing activity (Gao et al., 2006), that also plays an important role during bone regeneration, repairs and the induction of mesenchymal stem cells into osteocytes (Wang et al., 1990).
Native human BMP-2 is a homodimeric protein of identical monomer with a dominant beta-sheet structure and forms the cystine-knot assembly. Each monomer connected together with three intramolecular disulfide linkages and one interchain disulfide bridge to form an active dimer (Scheufler et al., 1999). The interface between the two monomers is stabilized by hydrophobic interactions and an intermolecular disulfide bond. For BMP-2 processing, a 396 amino acid precursor is proteolytically cleaved to yield the mature form of 114 amino acids (Hillger et al., 2005), and demonstrates the biological activity only in a dimeric form. The stabilization of disulfide bonds involves the protein interaction with transmembrane serine/threonine kinase receptors on osteogenic cells, leading to activate cell proliferation and differentiation of osteoblasts (Reddi, 2000). Owing to its osteoinductive capacity, BMP-2 renders a therapeutic protein for de novo bone formation in clinical use, which is utilized as an alternative to bone autografting during the healing of critical fractures, for spinal fusions or the treatment of bone and periodontal defects including dental implants (Gautschi et al., 2007; McKay et al., 2007; Bessa et al., 2008; Tang et al., 2009; Kimura et al., 2010; Shimono et al., 2010; Luo et al., 2012; Marques et al., 2015; Gomes-Ferreira et al., 2016; Poon et al., 2016; Herford, 2017; Gonzaga et al., 2019). However, isolation of hBMP-2 directly from bone is an economically unfeasible process because of very low yields in microgram quantities, the laborious purification process, and safety (potential health risks associated from allogeneic bone donor) (Kirker-Head, 2000).
Since its significance in therapeutic treatment; several heterologous expression systems have been published for recombinant hBMP-2. Heterologous production of constitutively active rhBMP-2 from mammalian systems like Chinese hamster ovary (CHO) cells (Wang et al., 1990; Israel et al., 1992) aids disulfide bond formation and protein folding, but has disadvantages of imperfect monomer processing, time intensive, expensive cultivation and poor yield with high variability (Jayapal et al., 2007). Similar problems occurred during the production of rhBMP-2 in virus infected insect cells (Maruoka et al., 1995) and transgenic plants (Gao et al., 2006). Expression of rhBMP-2 using bacterial systems may overcome those problems therefore it is an attractive alternative. Bacteria host cells are the most popular choice for the expression of human genes due to advantages such as rapid growth, low cost, high yield, and, that they can be manipulated for various purposes (Graumann and Premstaller, 2006). Production of biologically active rhBMPs through in vitro refolding of E. coli produced inclusion bodies have been reported ( Long et al., 2006; Sharapova et al., 2010; von Einem et al., 2010; Hyup Lee et al., 2011; Zhang et al., 2011; Pramesti et al., 2012; Retnoningrum et al., 2012; Rane et al., 2013). However, the refolding procedure was complicated, labour intensive, time consuming and required expensive reagents (Ruppert et al., 1996; Vallejo et al., 2002).
The purpose of this study is to develop an effective method to improve the production of an active rhBMP-2 as a soluble form without any refolding or solubilization steps. A 342-bp gene encoding mature human BMP-2, was isolated from a cDNA human osteoblast cell line hFOB 1.19 and expressed in SHuffle
E. coli, to promote a disulfide bond formation. The efficient method of expression and purification yields a soluble protein of rhBMP-2 with biological activities to induce the osteogenic properties in a human osteoblast cell.
MATERIALS AND METHODS
Cloning and construction of rhBMP-2
The nucleotide sequence of human BMP-2 gene (GenBank Accession no. NM_001200) from NCBI database was used as a reference gene. Human cDNA encoding 114 amino acids long mature form of BMP-2 (residue 283 to 396) was cloned by reverse transcription-PCR (RT-PCR). Total RNA was isolated from the cultured human fetal osteoblast cell line (hFOB 1.19 (ATCC® CRL-11372™, USA) by Nucleospin® RNA II (Machere-Nagel, Germany), and was reverse transcribed by RevertAid™ H Minus First Strand cDNA Synthesis Kit (Thermo scientific, USA) using oligo (dT)18 primer. The mature BMP-2 cDNA sequence was amplified by PCR using GoTaq® Green Master Mix (Promega, USA) with primers 5'-CAAGCCAAACACAAACAGCG-3' (20-mer) and 5'-GCGACACCCACAACCCTC-3' (18-mer). The PCR condition was the two minutes of pre-denaturation at 94 °C, 35 cycles of 45 sec at 94 °C, 1 min at 55 °C, 1 min at 72 °C, and 5 minutes for post-elongation at 72 °C. For cloning purposes, the sequence encoding EcoR I, Nco I, six histidine residues and recognition sequence of factor Xa protease was introduced to 5¢ end (forward primer) whilst termination codon and BamH I restriction sites to the 3¢ end of hBMP-2 gene (reverse primer). The forward primer had the sequence of 5'-GAATTCGGCCATGGCTCATCACCATCACCATCACATCGAAGGGCGCCAAGCCAAACACAA-3' (60-mer) and the reverse primer had the sequence of 5'-CGCAAGCTTGGATCCTTAGCGACACCCACAACC-3' (33-mer). The PCR reaction was performed with the following conditions, five minutes of pre-denaturation at 95 °C, 30 cycles of 30 sec at 95 °C, 30 sec at 58 °C, and 30 sec at 72 °C, and 5 minutes for post-elongation at 72 °C. The amplified product was ligated into CloneJETTM PCR Cloning kit (Thermo scientific, USA) and transformed into competent E. coli XL1-Blue. The digested EcoR I/BamH I fragment was purified using a gel extraction kit (QIAGEN, Germany) and sub cloned into the corresponding sites of pEZZ18 Protein A Gene Fusion Vector (GE Healthcare, USA). The recombinant clones were randomly screened by colony PCR, analyzed with EcoR I/BamH I restriction digestion, and finally verified the insert DNA sequence by automated DNA sequencing (First BASE Laboratories, Malaysia). The resulting nucleotide sequences were analyzed using a Blast Similarity search tool on NCBI database. The expression vector pEZZ18 harbouring hBMP-2 gene was transformed into SHuffle® Express Competent E. coli (New England Biolabs, USA) for protein expression.
Soluble expression and purification
The pEZZ18-hBMP-2 in SHuffle Express E. coli was cultured at 30°C overnight in Luria-Bertani (LB) medium supplemented with 100µg/ml ampicillin. The fresh culture was inoculated with 1%inoculum into the LB media, then grown at 30°C at 180 rpm agitation until the OD600 reached 0.5-0.6. Thereafter, rhBMP-2 expression was induced by adding isopropyl b-D-thiogalactopyranoside (IPTG) at a final concentration of 1 mM and further cultured for 18 h at 30°C. The bacterial cells were harvested by centrifugation at 2,300xg for 5 min at 4°C and resuspended in a lysis buffer (50mM NaH2PO4, pH 8.0, 300mM NaCl) containing protease inhibitor cocktails (Amresco, USA), then the cells were disrupted using a sonicator. After removing the cell debris by centrifugation at 15,300xg for 25 min at 4°C, the clear supernatant was applied to pre-equilibrated IgG Sepharose 6 Fast Flow resin (GE Healthcare, USA) and then incubated at 4°C with gentle agitation overnight. The column was washed with the binding buffer (TBST; 20mM Tris-HCl, pH 7.5, 150mM NaCl, 0.05%Tween 20) and followed by a washing buffer (TBST containing 0.6M NaCl) and finally washed with TBS (20mM Tris-HCl, pH 7.5, 150mM NaCl). The bound proteins were eluted with 50mM ammonium acetate with 2M MgCl2, pH 5.0. The eluted fractions after measurement at OD280 were pooled, concentrated and exchanged with 20mM Tris-HCl, pH 8.0 using Amicon ultra-4 MW cut off 10 kDa (Millipore, USA). The protein was kept in aliquots with 20mM Tris-HCl, pH 8.0 buffer containing 20%glycerol at -80°C, and the concentrations were determined by Bradford protein assay with BSA as a standard.
SDS-PAGE and Western blot analysis
The proteins were resolved on 12% SDS polyacrylamide gel, then the bands were either stained by Coomassie Brilliant Blue R-250 or analyzed by Western blot. For Western blotting, proteins were electro-transferred to the PVDF membrane using semi-dry blotting apparatus (Biorad) at a constant voltage of 20 V for 20 minutes. It was then blocked with 5% non-fat milk in TBST for 1 hour at room temperature and incubated overnight at 4°C with rabbit anti-hBMP-2 polyclonal antibody (Biovision, USA) at dilution 1:2000. The membrane was washed 3 times with a TBST buffer and then incubated with a goat anti-rabbit horseradish peroxidase (Merck, USA) at 1:4000 dilution for 1 hour at room temperature. After washing with TBST, the immune-blots were visualized by LuminataÔ western chemiluminescent HRP substrate (Merck, USA).
Protein identification by LC-MS/MS mass spectrometry
The protein band was excised from a Coomassie Brilliant Blue-stained gel after separating from 12% SDS-PAGE gel, reduced with DTT, alkylated with iodoacetamide and digested with trypsin. The digested peptide solutions were analyzed with Impact II UHR-TOF MS system (Bruker Daltonics, USA) coupled to a nanoLC system: UltiMate 3000 LC system (Thermo Fisher Scientific, USA). Peptides were separated on a nanocolumn (PepSwift monolithic column 100 mm i.d. x 50 mm). Eluent A was 0.1% formic acid and eluent B was 80% acetonitrile in 0.1% formic acid. Peptide separation was achieved with a linear gradient from 10% to 45% B for 8.5 min at a flow rate of 1 ml/min, including a regeneration step at 90% B and an equilibration step at 1% B, one run took 20 min. Peptide fragment mass spectra were acquired in data-dependent AutoMS (2) mode with selecting most abundant precursor ions in a 3 second cycle for fragmentation. The mass range of the MS scan was set to extend from 150 to 2200 m/z. The MS/MS data was searched against the local database using multiple-search-engines available in SearchGUI and the obtained proteomics identification results were interpreted by Peptide Shaker (Vaudel et al., 2011; Vaudel et al., 2015) .
Cell proliferation assay
The effect of rhBMP-2 on cell proliferation was performed with PrestoBlueÔ cell viability reagent (Invitrogen, USA). The hFOB 1.19 cells were seeded at a density of 1 x 104 cells/well in the 96-well black plate and maintained in 1:1 mixture of phenol red-free DMEM/Ham¢s F-12 medium (Sigma-Aldrich, USA) supplemented with 10%fetal bovine serum (FBS), 100U/ml penicillin, 100 mg/ml streptomycin, 0.3mg/ml G418, 1.2 g/L NaHCO3 and 2.5 mM sodium pyruvate at 37oC with 5%CO2. After 24 h of incubation, the cells were washed with PBS buffer and changed with fresh medium containing 0.5% FBS with rhBMP-2 at different amounts (3.125 to 50 ng/ml) or E. coli-derived hBMP-2 (R&D Systems, USA) as a positive control. After 4 h of culture, the cell growth was determined by PrestoBlueÔ cell viability assay which based on a resazurin-based method of the reducing ability of living cells to quantitatively measure cell proliferation. The fluorescence signal was measured at excitation 530 nm and emission 590 nm using multi-mode microplate reader (SynergyÔ HT, BioTek, USA).
Alkaline phosphatase staining
hFOB 1.19 cells were plated at 6 x 104 cells/well in a 24-well plate and cultured until reached 70-80% confluence in a completed medium. Cells were treated with rhBMP-2 or E. coli-derived hBMP-2 at concentrations of 25 and 50 ng/ml in a medium containing 2% FBS. Cells were continuously cultured at 37oC with 5%CO2 for 48 hours. The treated cells were carefully washed with PBS, fixed with 10% neutral buffered formalin for 10 minutes and then rinsed with PBS. After the fixing step, the cells were stained with BCIP-NBT solution (Amresco, USA) and incubated at room temperature in the dark for 1 hour. The reaction was stopped by removing the substrate solution and washed with PBS. Photographs were taken to evaluate the staining results using an inverted microscope with digital imager (IX71, Olympus, Japan).
Alkaline phosphatase activity assay
The quantitative analysis of rhBMP-2 on the induction of alkaline phosphatase (ALP) activity in hFOB 1.19 was determined by the modified colorimetric method (Wanachewin et al., 2015). The cells were seeded as previously described and replaced with rhBMP-2 or E. coli-derived hBMP-2 (R&D Systems, USA) in a medium containing 2% FBS. After treatment for 5 days, the cells were washed and the attached cells were scraped off. The cells were lysed with 100 ml phosphatase buffer followed by sonication and the protein concentration was determined using Bradford assay. ALP activity assay was performed in a 96-well plate by adding the cell lysate with 150mM alkaline buffer solution (Sigma-Aldrich, USA). The reaction was initiated by adding 1 mg/ml phosphatase substrate solution (Sigma-Aldrich, USA) and incubated at 37°C for 60 min. The reaction was stopped by 0.1N NaOH when the color intensity was measured at 405 nm using a microplate reader. The alkaline phosphatase activity was quantified by the production of p-nitrophenol (mM) product per mg protein per minute and compared as a percentage with the untreated control (100%).
Gene expression by quantitative real-time PCR
The hFOB 1.19 cells were cultured in 6-well plates and exposed with rhBMP-2 at 25 and 50 ng/ml in media containing 0.5% FBS. After 12-h treatment, the total RNA was extracted by RibozolTM RNA extraction reagents (Amresco, USA) and the concentration was determined by a NanoDropÔ 2000 spectrophotometer. One microgram of total RNA was then reverse-transcribed to cDNA using the RevertAidÔ First Strand cDNA Synthesis kit (Thermo scientific, USA). A real-time quantitative polymerase chain reaction was performed in triplicate samples by a LightCycler® 96 Real-Time PCR System using Fast Start Essential DNA Green Master (Roche, Switzerland) following the manufacturer’s instructions. Primer sequences were summarized in Table 1. The quantification cycle (Cq) values were calculated using Light Cycle 96 SW 1.1. Relative expression levels for each primer set were normalized to the expression of GAPDH as a reference gene. N-fold difference expression (relative to control) was calculated using the 2-DDCq method (Livak and Schmittgen, 2001). Mean fold difference was calculated and represented as ± standard deviation.
Table 1. Primers used for gene expression by real-time qRT-PCR.
|
Gene |
Primer sequence (5¢®3¢) Forward (F); Reverse (R) |
Accession number |
|
|
ALP |
F : CATGGCTTTGGGCAGAAGGA R : CTAGCCCCAAAAAGAGTTGCAA |
NM_001114107.4
|
|
|
COL1 |
F : AGCCGCTTCACCTACAGC R : TTTTGTATTCAATCACTGTCTTGCC |
NM_000088.3 |
|
|
BMP-2 |
F : GCTGTCTTCTAGCGTTGCTG R : CTGTTTCAGGCCGAACATGC |
NM_001200.3 |
|
|
Runx2 |
F : GCCTTCAAGGTGGTAGCCC R : CGTTACCCGCCATGACAGTA |
NM_001024630.3
|
|
|
OPN |
F : ATGGCCGAGGTGATAGTGTG R : TCAGGGTACTGGATGTCAGG |
NM_000582.2
|
|
|
GAPDH |
F : GAAGGTGAAGGTCGGAGTC R : GAAGATGGTGATGGGATTTC |
NM_002046.5 |
|
Statistical Analysis
All data were presented as the mean ± SD. Comparison among groups were performed by one-way ANOVA analysis using SPSS statistic version 17.0 followed by Tukey's t-test. Differences were considered significant at a value of P < 0.05 for all tests.
RESULTS
Cloning and construction of rhBMP-2
A full-length of gene encoding for mature hBMP-2 was achieved from the human fetal osteoblast cell line (hFOB1.19) by RT-PCR. Human BMP-2 cDNA was amplified by PCR and gave a single band at 342 bp of mature hBMP-2.
In order to facilitate the cloning step, the EcoR I and BamH I restriction sites were added at its 5' and 3'ends, respectively, whereas the histidine 6 amino acids and factor Xa were fused at N-terminus to facilitate the purification step. The amplified product at 406 bp in length was observed and ligated into pJET1.2 plasmid vector. The coding region of the mature hBMP-2 was subcloned into the corresponding sites of the expression vector pEZZ18 to generate pEZZ18-hBMP-2 (Figure 1) and characterized by PCR. Both plasmids were analyzed by the double restriction enzymes with EcoR I and BamH I, showing the hBMP-2 gene fragment. The nucleotide sequence of the insert fragment was a perfect match (100%) with the mature hBMP-2 gene using nucleotide blast search on NCBI database, confirming that the recombinant clones harbored the hBMP-2 gene. Translated amino acid sequences of protein product showed the fusion protein rhBMP-2 (302 amino acids) containing ZZ peptide, histidine 6 amino acids, factor Xa cleavage region and mature hBMP-2 (Figure 2).

Figure 1. The pEZZ18-hBMP-2 construct.
MMTLQIHTGGINLKKKNIYSIRKLGVGIASVTLGTLLISGGVTPAANAAQ
HDEAVDNKFNKEQQNAFYEILHLPNLNEEQRNAFIQSLKDDPSQSANLLAE
AKKLNDAQAPKVDNKFNKEQQNAFYEILHLPNLNEEQRNAFIQSLKDDPSQ
SANLLAEAKKLNDAQAPKVDANSAMAHHHHHHIEGRQAKHKQRKRLK
SSCKRHPLYVDFSDVGWNDWIVAPPGYHAFYCHGECPFPLADHLNS
TNHAIVQTLVNSVNSKIPKACCVPTELSAISMLYLDENEKVVLKNYQ
DMVVEGCGCR
Figure 2. Translated amino acid sequence of rhBMP-2 fusion protein. Amino acids in regular letters belong to tag fragment while those in bold letters correspond to mature human BMP-2 protein. ZZ peptide is shown in italic letters; followed by histidine 6 amino acids and factor Xa cleavage region, respectively. Underlined letters are residues identified from LC-MS/MS analysis.
Expression and purification of rhBMP-2
The initial investigation for expression of rhBMP-2 at 37°C observed that the resultant protein product consisted of only inactive inclusion bodies in the crude extract of the transformed bacteria. Induction with IPTG at 30°C for 18 h yielded a soluble protein in the crude extract, whereas some inclusion bodies were remained. The rhBMP-2 from the soluble fraction was purified with a single step by affinity chromatography using IgG Sepharose 6 Fast Flow. The eluted proteins appeared in a single band with the migrated protein at about 67 kDa with > 95% purity (Figure 3A) on the coomassie-stained gel. Western blot analysis with polyclonal antibodies against hBMP-2 detected a purified protein product at about 67 kDa, confirming that the purified protein was rhBMP-2 (Figure 3B). The overall yield of purified rhBMP-2 was approximately 4 mg per liter of culture medium with 11.3% yield. Theoretical isoelectric point of fusion protein rhBMP-2 calculated on Expasy based on a deduced amino acid sequence, a web-based program was 7.26, whereas the molecular mass on monomer was 33.7 kDa.
(A) (B)
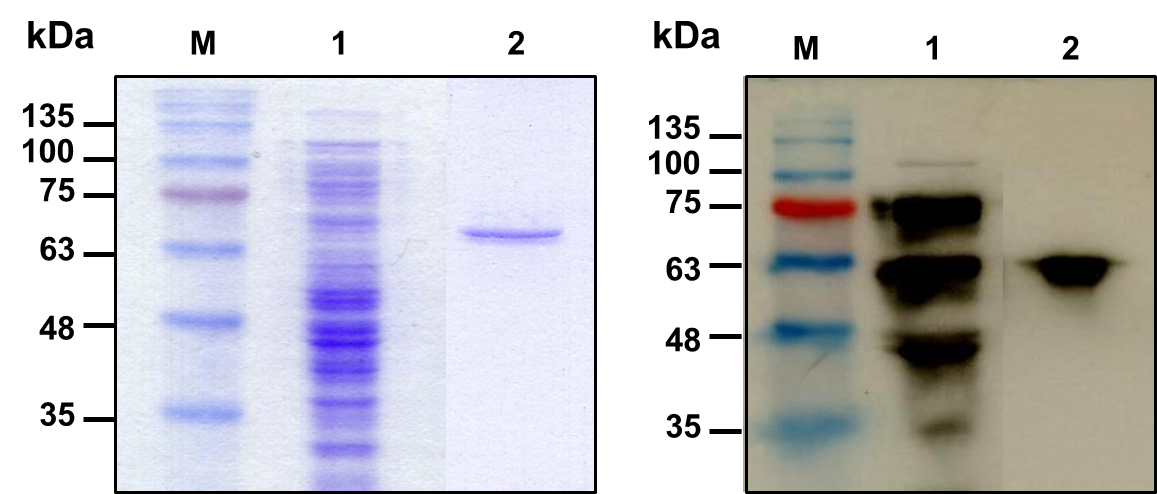
Figure 3. Analysis of expression and purification for rhBMP-2. (A) Coomassie-stained 12% reducing SDS-PAGE of purified protein samples. (B) Western blot analysis using polyclonal antibody to human BMP-2 developed by chemiluminescent substrate. Lane M, prestained protein marker; lane 1, supernatant fraction from IPTG-induced SHuffle expressed E. coli harboring pEZZ18-hBMP-2; lane 2, purified rhBMP-2 after IgG Sepharose 6 Fast Flow purification.
Confirmation of rhBMP-2 using mass spectrometry
Purified protein was analyzed by LC-MS/MS and further identified using multiple proteomics identification search engines, including OMSSA, MariMatch X!Tandem, Andromeda, Comet and more which were available in SearchGUI. A Peptide Shaker was used to increase the confidence and sensitivity of combined results from multiple search engines. Six tryptic peptides were identified and assigned to the sequence corresponding to the coverage of 31.79% as summarized in Table 2. These results verified that the purified protein was rhBMP-2.
Table 2. Results of LC-MS/MS analysis of purified rhBMP-2 matched with human bone morphogenetic protein-2.
|
Amino acid residue |
Calculated mass (m/z) |
Charge |
Peptide sequence |
Modification |
|
||||
|
1-16 |
615.98 |
+3 |
MMTLQIHTGGINLKKK |
Oxidation at M |
|||||
|
82-89 |
461.26 |
+2 |
NAFIQSLK |
- |
|||||
|
139-147 |
461.26 |
+2 |
NAFIQSLK |
- |
|||||
|
163-188 |
485.87 |
+6 |
LNDAQAPKVDANSAMAHHHHHHIEGR |
Oxidation at M |
|||||
|
265-289 |
483.95 |
+6 |
ACCVPTELSAISMLYLDENEKVVLK |
Oxidation at M |
|||||
|
290-302 |
802.89 |
+2 |
NYQDMVVEGCGCR |
Oxidation at M |
|||||
The effect of rhBMP-2 on cell proliferation
The biological activities to induce the osteogenic properties of rhBMP-2 were examined in vitro using human osteoblast cells (hFOB 1.19). The effect of rhBMP-2 on cell proliferation is shown in Figure 4 based on Prestoblue™cell viability assay. The hFOB 1.19 cells were cultured and treated with different rhBMP-2 concentrations for 4 hours. As expected, the rhBMP-2 significantly stimulated cell growth increasing from 100% to 229% from 3.125 to 50 ng/ml in a dose dependent manner. Similar effects were observed when treated with commercial E. coli-derived rhBMP-2. Therefore, the purified rhBMP-2 was functionally active in inducing cell proliferation on human osteoblast cells.
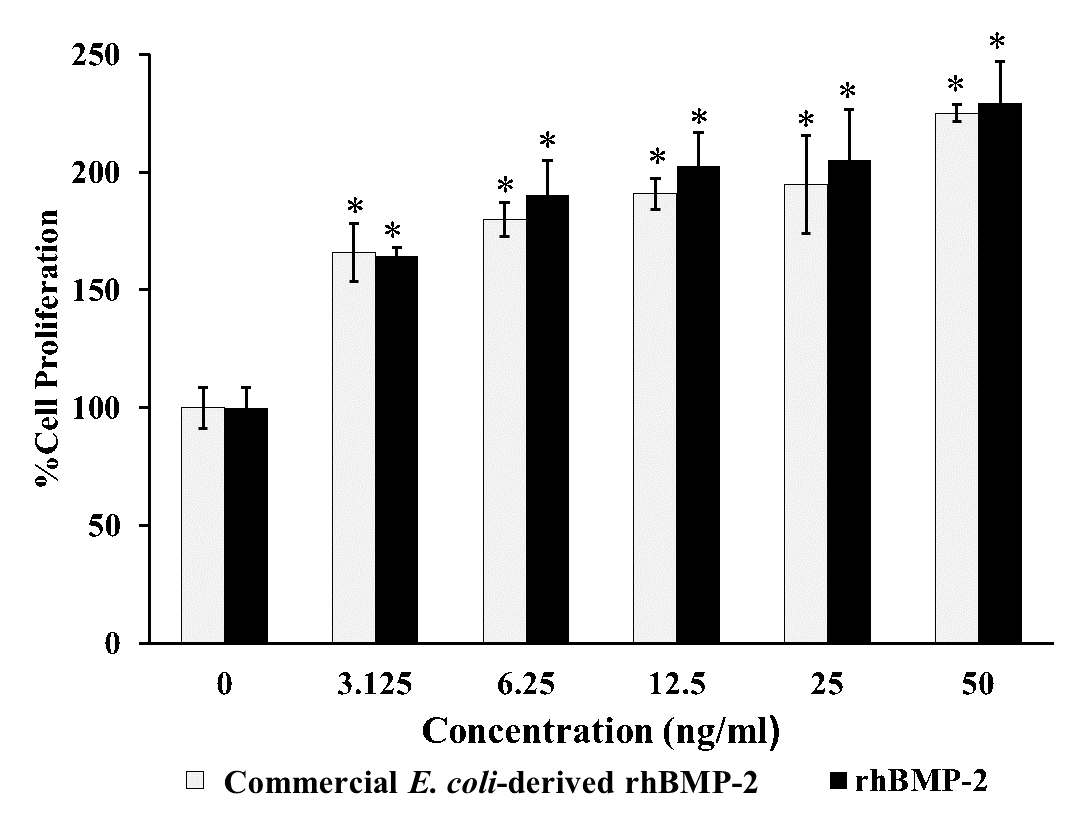
Figure 4. Cell proliferation of rhBMP-2 on hFOB 1.19 cells. Graph showing the effect of purified rhBMP-2 on proliferation by PrestoBlue™ reagent compared with commercial E. coli-derived rhBMP-2 (R&D Systems) at 4-h treatment with the various rhBMP-2 concentration of 3.125, 6.25, 12.5, 25 and 50 ng/ml, respectively. All data are shown as mean ± S.D. of triplicate experiments. Statistical analysis was analyzed by one-way ANOVA by SPSS followed by Tukey's t-test.
Note: *denoted for P-value < 0.05 was statistically significant.
The effect of rhBMP-2 on cell differentiation
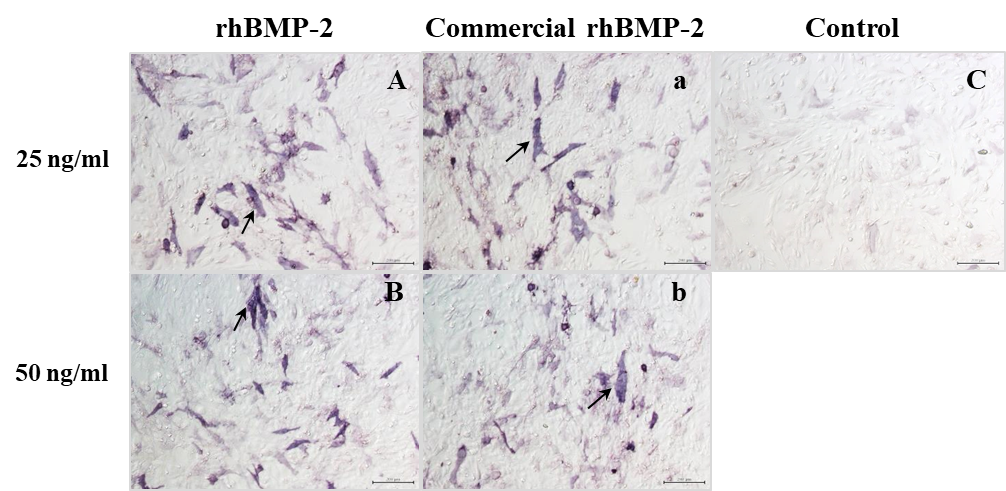
ALP activity is an early biomarker of osteogenesis and quantitatively estimated the degree of osteoblast differentiation. The potential effect of rhBMP-2 to induce the production of alkaline phosphatase was processed by staining, using BCIP/NBT as a substrate. The staining results after rhBMP-2 treatment for 48 hours appeared the positive reactivity (blue-purple colour) at 25 and 50 ng/ml (Figure 5A). After 5 days of treatment, the ALP activity of rhBMP-2 at 25 and 50 ng/ml significantly increased its activity at 170% and 210% compared to the untreated control (100%control) (Figure 5B). Similar results were shown after treatment with commercial E. coli-derived rhBMP-2 (Figure 5A and B). Thus, rhBMP-2 had the biological activities necessary to stimulate osteoblast differentiation.
(A)
(B)
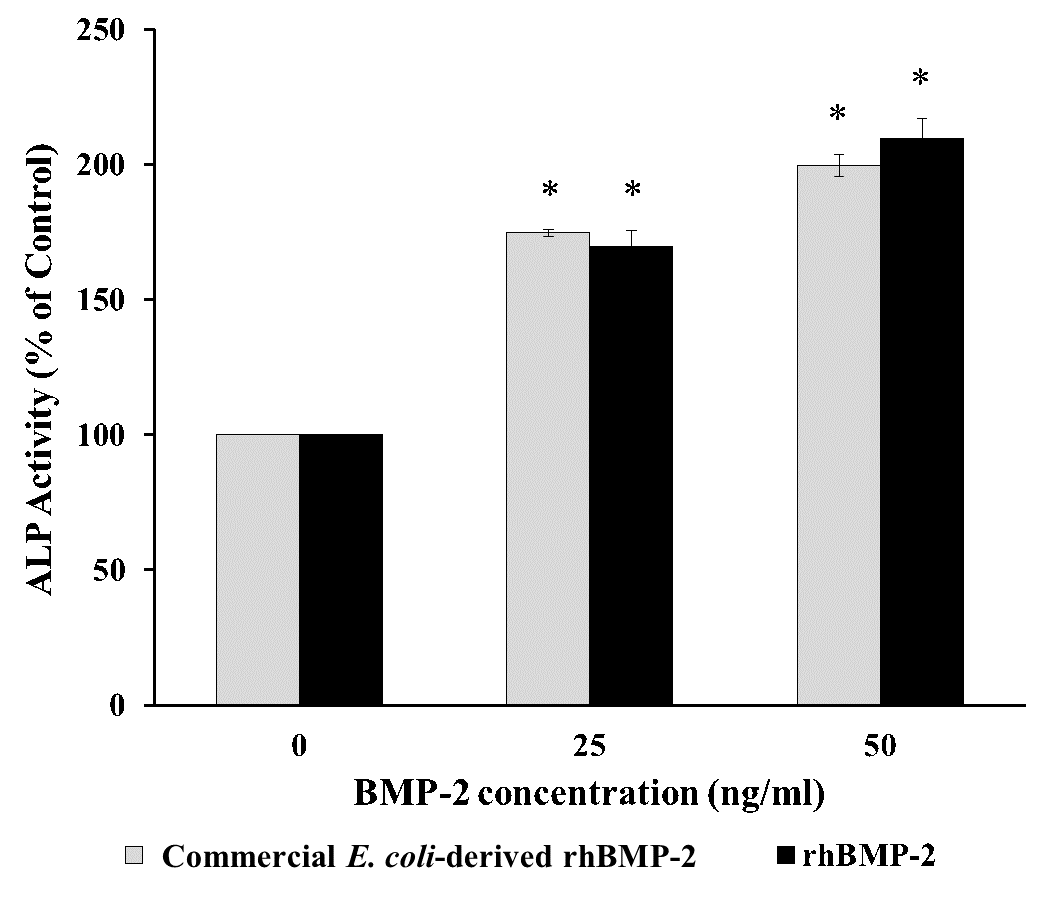
Figure 5. Alkaline phosphatase detection on hFOB 1.19 after treatment with rhBMP-2. (A) Alkaline phosphatase staining using NBT-BCIP substrate solution. A and B, treatment with purified rhBMP-2; a and b; treatment with commercial E. coli-derived rhBMP-2; C, no treatment as a control. A, a = 25 ng/ml; B, b = 50 ng/ml. Magnification = 10x, Bar = 200 mm. Blue-purple color of positive alkaline phosphatase staining was pointed by black arrow. (B) Alkaline phosphatase activity after treatment with purified rhBMP-2 for 5 days. Data represent mean ± standard deviation (n = 3), *P < 0.05 compared to control.
Gene expression analysis of rhBMP-2
To confirm the functional activity of rhBMP-2 on the osteoblast differentiation process, we examined the degree of mRNA expression for genes associated with osteoblast differentiation, alkaline phosphatase (ALP), type I collagen (COL1), bone morphogenetic protein-2 (BMP-2), runt related protein 2 (Runx2) and osteopontin (OPN). Relative quantitation of Real-Time PCR results is shown in Fig. 6 where the mRNA transcript of ALP, COL1 and BMP-2 in the cells was stimulated by rhBMP-2 at 25 ng/ml (Figure 6A). The mRNA expression levels of ALP, COL1 and BMP-2 were increased to 4.3-fold, 6.7-fold and 4.2-fold relative to control, respectively. The up-regulated gene expression was also significantly increased to Runx2 transcription factor and OPN was significantly increased to 2.2-fold and 2.3-fold at 50 ng/ml treatment, respectively (Figure 6B). Therefore, the rhBMP-2 could significantly increase the expression level of osteoblastogenic genes in human osteoblast cells during osteoblast differentiation.
(A) (B)
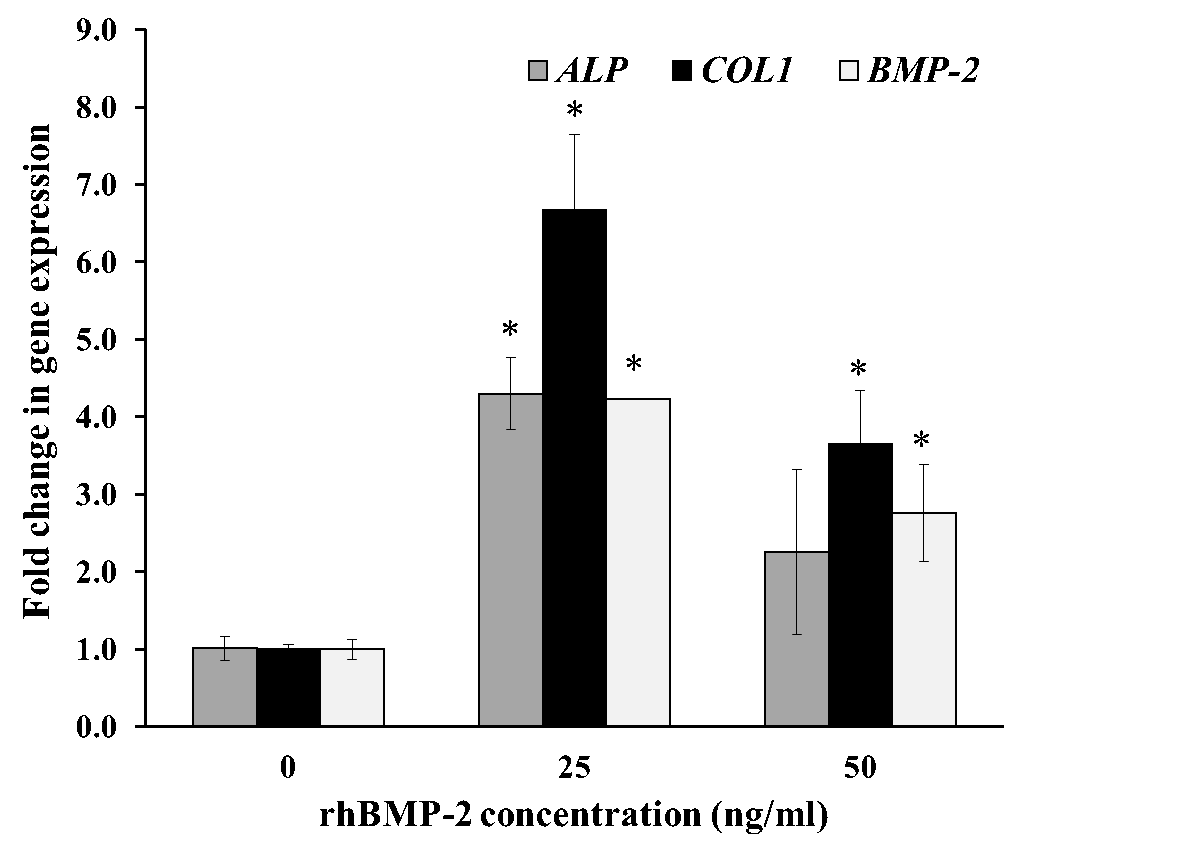
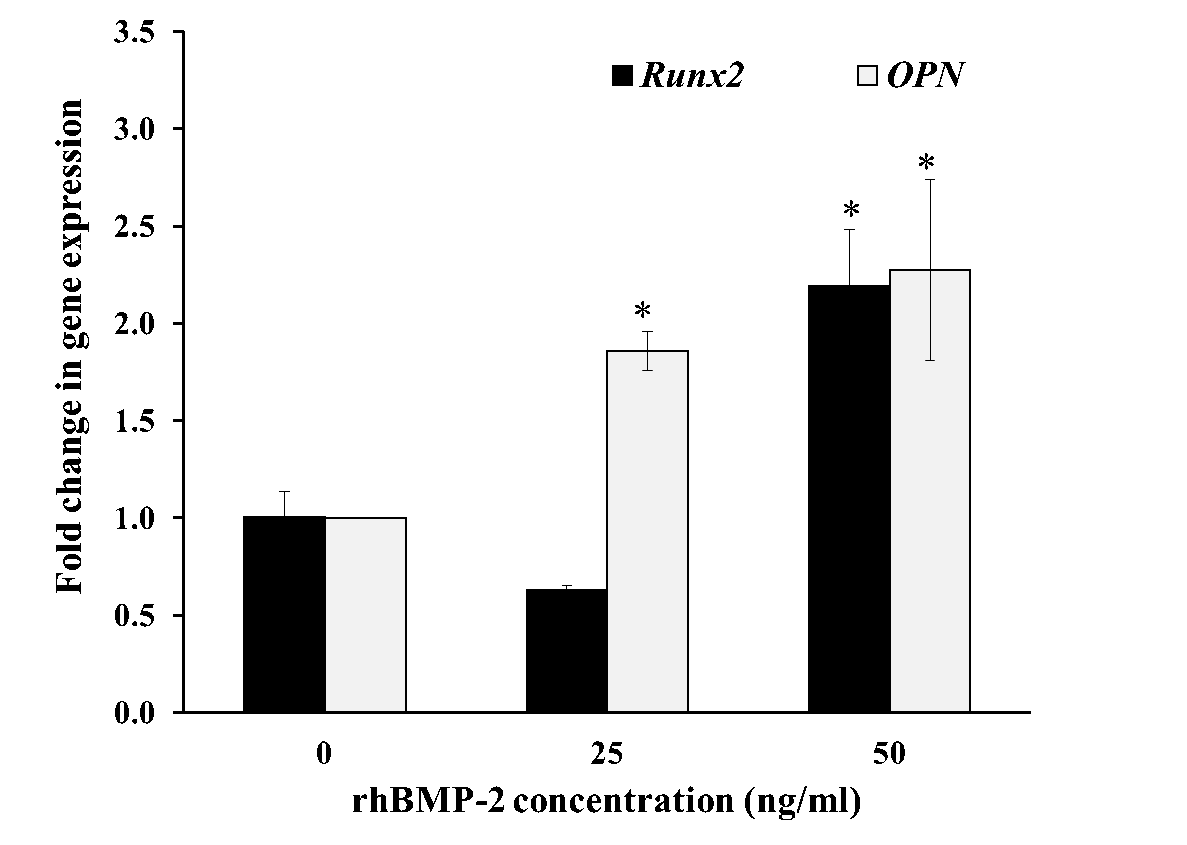
Figure 6. Gene expression analysis by qRT-PCR with rhBMP-2 treatment. hFOB 1.19 cells were cultured for 12 h at 25 and 50 ng/ml purified rhBMP-2. Fold change in mRNA expression level are relative to GAPDH and normalized in the untreated control groups. Relative gene expression of ALP, COL1, BMP-2 (A), and Runx2, OPN (B) shows in the figure. Control group was taken as the calibrator. All data are shown as mean ± S.D. of triplicate experiments. Statistical significance of differences was assessed with one-way ANOVA by SPSS followed by Tukey's t-test. P-value < 0.05 (*) was assumed statistically significant.
DISCUSSION
Human BMP-2 is a crucial growth factor for osteoinductive and osteogenesis processes of bone development and regeneration (Poon et al., 2016). Native hBMP-2 is a disulfide bonded homodimeric protein which is needed for biological functioning and is accompanied by a posttranslational modification of N-glycosylation (Wang et al., 1990). The absence of carbohydrate residues does not affect the biological function of BMP-2 (Kubler et al., 1998). Previous studies (Long et al., 2006; Sharapova et al., 2010; von Einem et al., 2010; Hyup Lee et al., 2011; Zhang et al., 2011; Pramesti et al., 2012; Rane et al., 2013; Nasrabadi et al., 2018; Quaas et al., 2018) have expressed rhBMP-2 from a prokaryotic host in the form of inclusion bodies by the pET expression system under the control of T7 promoter using E. coli BL21(DE3), OrigamiTM E. coli or E. coli SHuffle T7 Express strain. Nevertheless, these bacterial expression systems are limited in their ability to perform post-translational modifications and facilitate disulfide bond formation, resulting in eukaryotic proteins that are improperly folded with the formation of inactive inclusion bodies. The misfolded protein needs further in vitro refolding in order to regain its active structure. The complex nature of this process is time-consuming, labor intensive and dependent upon various variables for optimal conditions. Inclusion bodies may be formed due to there being no disulfide bond formation in the reducing state of the cytosol in E. coli.
In the current study, we have investigated an E. coli-based expression system to produce a soluble and functionally active rhBMP-2 protein using the human osteoblast cell line (hFOB 1.19) as an in vitro biological testing. Our strategy for improvement of the soluble expression of rhBMP-2 was circumvented by changing the expression conditions which included the use of a weaker promoter, the co-expression with fusion protein, lowering the growth temperature and using a specific host strain allowing the formation of disulfide bridges in E. coli host cells. The rhBMP-2 was successfully expressed as soluble protein after induction by IPTG with the pEZZ18 expression vector by using the bacterial host strain SHuffle E. coli. The fusion partner with ZZ peptide at its N-terminal was selected for helping the protein folding and aided in the purification step. The active rhBMP-2 was expressed as a soluble protein under the control of lacUV5 and a protein A promotor at a lower temperature (30°C). These results indicated that that type of promoter and a lower culture temperature increased the soluble expression due to a slow rate of protein synthesis (Rosano and Ceccarelli, 2014). Nevertheless, the rhBMP-2 protein was expressed in low amount of total cell protein, which may be circumvent this problem by reducing temperature (<30°C) or co-expression with chaperone proteins (Martínez-Alonso, 2010). Furthermore, the E. coli SHuffle strain produced the correct folding of active rhBMP-2 protein within its cytoplasm. This agreement with the information that this strain is based on the deletions of the genes for glutaredoxin reductase and thioredoxin reductase, which allows disulfide bonds to form in the cytoplasm (Lobstein et al., 2012). It also expresses the cytoplasmic disulfide bond isomerase DsbC which helps the oxidizing cytoplasmic environment and its role in assisting the formation of correctly folded multi-disulfide bonded proteins (Lobstein et al., 2012). The advantages of expression as a soluble protein with an active protein in the E. coli system include inexpensive, less time and no refolding and solubilization steps for protein purification. Recombinant hBMP-2 protein was employed in a single step by affinity chromatography with IgG Sepharose 6 Fast Flow column. This data suggested that rhBMP-2 fusion with ZZ peptide was enriched with the IgG Sepharose 6 Fast Flow column which is tightly bound with the IgG binding domain of protein A. The storage of rhBMP-2 was held at the alkaline buffer, pH 8.0 with 20%glycerol. At pH 8.0, the biological function of the proteins is maintained, as reported previously on the usage of rhBMP-2 in alkaline solution (Sharapova et al., 2010). The molecular mass at approximately 67 kDa was detected by both SDS-PAGE and western blot analysis, suggested that the rhBMP-2 might be a homodimeric form which composed of a monomeric 33.7 kDa hold together. The molecular mass of rhBMP-2 monomeric form (33.7 kDa) is larger than native mature hBMP-2 (12.9 kDa) because it composed of the protein fragment containing ZZ peptide, histidine 6 amino acids, factor Xa cleavage region (20.8 kDa) and mature hBMP-2 (12.9 kDa). However, the purified rhBMP-2 dimer was insensitive to SDS, heat and beta-mercaptoethanol, suggesting that this dimerization creates a hydrophobic core between the monomers that stabilizes the molecule (Scheufler et al., 1999).
Since the biological activity of rhBMP-2 is the key indicator for successful protein production, we demonstrated the osteogenic effect of E.coli-derived rhBMP-2 using human fetal osteoblast cells, not rodent cells, such as C2C12 cells. Most studies evaluated the osteogenic activity of E.coli-derived rhBMP-2 using the C2C12 cell line (Fung et al., 2019). Proliferation of hFOB 1.19 cells was promoted by increasing the rhBMP-2 concentration, indicating the rhBMP-2 contains the ability to stimulate the proliferation of human osteoblast cells. This result agreed well with high biological activity of BMP-2 in nanogram level reported previuosly (Long et al., 2006). Alkaline phosphatase is a key enzyme in the osteogenic differentiation process and its activity increases with the enhancement of osteoblast differentiation. The increasing of ALP activity indicated that rhBMP-2 was able to induce osteoblast development from the pre-osteoblast to the extracellular matrix maturation stage. The purified rhBMP-2 has a similar level of activity to commercially available rhBMP-2, indicating that our rhBMP-2 has the functional activity as same as a commercial product. During the bone formation process, increased expression of specific genes in playing roles in the extracellular matrix formation and mineralization was reported (Kirkham and Cartmell, 2007). The up-regulation of five osteoblast-associated genes including ALP, COL1, BMP-2, Runx2 and OPN indicated that rhBMP-2 could stimulate the osteoblast differentiation process. ALP and collagen type I are the early indicators of osteoblastic differentiation. Collagen type I is an important component of bone extra-cellular matrix, forming connections with cell surface integrins and other ECM proteins. BMP-2 is an osteoinductive protein, that is capable of inducing bone formation and healing, and it also promotes cell proliferation and differentiation towards the osteogenic pathway. Runx2 is an important transcriptional factor on osteogenic differentiation and controls bone formation. OPN is one of the noncollagenous proteins involved in the bone matrix organization. The osteoinductive properties of purified rhBMP-2 suggest that it might be a homodimeric protein of two monomers with identical subunits held together due to the biological function of the dimer (Long et al., 2006; Sharapova et al., 2010; von Einem et al., 2010; Hyup Lee et al., 2011; Zhang et al., 2011; Pramesti et al., 2012; Rane et al., 2013). This rhBMP-2 demonstrated in vitro biological activity capable of inducing osteogenic abilities which coincided with positive control commercial preparation of R&D research products. In our study, the purification process exposure to acidic conditions may be deactivated endotoxin (Ribeiro et al., 2010). Furthermore, the biological function of rhBMP-2 could stimulate the hFOB1.19 cells, suggesting that no or very low endotoxin contamination contained in our purified protein. To address this question, the quantitative measurement of bacterial endotoxin may be further verified using endotoxin detection assay.
CONCLUSION
The E. coli-based system to produce soluble rhBMP-2 with biological activity to stimulate the osteoinductive properties on a human osteoblast cell was established. This optimized expression and a single step purification are beneficial in the rapid production of active rhBMP-2 from E. coli without any refolding or solubilization steps from inclusion bodies. This information has potential applications for bone regenerative research, therapeutic intervention in bone disease and the production of other homodimeric proteins.
ACKNOWLEDGEMENTS
This project was supported by grants from the National Research Council of Thailand (Grant No. R2557B031) and the TRF-CHE Research Grant for New Scholar (Grant No. MRG5180032), Thailand Research Fund (TRF) and Commission on Higher Education (CHE), Ministry of Education, Thailand (to W.K.). In addition, we thank the Department of Biochemistry, Faculty of Medical Science, Naresuan University for use of its facilities. This work also supported by Thailand Excellence Center for Tissue Engineering and Stem Cells, Department of Biochemistry, Faculty of Medicine, Chiang Mai University, Chiang Mai, Thailand (to P.K.). Finally, we thank Kevin Mark Roebl, Division of International Affairs and Language Development, Naresuan University for proofreading and editing this manuscript.
REFERENCES
Balemans, W., and Van Hul, W. 2002. Extracellular regulation of BMP signaling in vertebrates: a cocktail of modulators. Development Biology. 250(2): 231-250. https://doi.org/10.1006/dbio.2002.0779
Bessa, P.C., Casal, M., and Reis, R.L. 2008. Bone morphogenetic proteins in tissue engineering: The road from the laboratory to the clinic, part I (basic concepts). Journal of Tissue Engineering and Regenerative Medicine. 2(1):1-13. https://doi.org/10.1002/term.63
Du, Y., and Yip, H. 2010. Effects of bone morphogenetic protein 2 on Id expression and neuroblastoma cell differentiation. Differentiation. 79(2): 84-92. https://doi.org/doi:10.1016/j.diff.2009.10.003
Fung, S.L., Wu, X., Maceren, J.P., Mao, Y., and Kohn, J. 2019. In Vitro Evaluation of recombinant bone morphogenetic protein-2 bioactivity for regenerative medicine. Tissue Engineering Part C : Methods. 25(9): 553-559. https://doi.org/ 10.1089/ten.TEC.2019.0156
Gao, Y., Suo, G.L., Han, J., He, Z.Q., Yao, W., and Dai, J.W. 2006. Expression of human BMP-2 gene in different tissues of tobacco plants. Acta Genetica Sinica. 33(1): 56-62. https://doi.org/ 10.1016/s0379-4172(06)60009-7
Gautschi, O.P., Frey, S.P., and Zellweger, R. 2007. Bone morphogenetic proteins in clinical applications. ANZ Journal of Surgery. 77(8): 626-631. https://doi.org/10.1111/j.1445-2197.2007.04175.x
Gomes-Ferreira, P.H., Okamoto, R., Ferreira, S., De Oliveira, D., Momesso, G.A., and Faverani, L.P. 2016. Scientific evidence on the use of recombinant human bone morphogenetic protein-2 (rhBMP-2) in oral and maxillofacial surgery. Oral Maxillofac Surgery. 20(3): 223-32. https://doi.org/ 10.1007/s10006-016-0563-4.
Gonzaga, M.G., Dos Santos Kotake, B.G., de Figueiredo, F.A.T., Feldman, S., Ervolino, E., Dos Santos, M.C.G., Issa, J.P.M. 2019. Effectiveness of rhBMP‐2 association to autogenous, allogeneic, and heterologous bone grafts. Microscopy Research and Technique. 82(6): 689-695. https://doi.org/10.1002/jemt.23215
Graumann, K., and Premstaller, A. 2006. Manufacturing of recombinant therapeutic proteins in microbial systems. Biotechnology Journal. 1(2): 164-186. https://doi.org/ 10.1002/biot.200500051
Herford, A.S. 2017. The use of recombinant human bone morphogenetic protein-2 (rhBMP-2) in maxillofacial trauma. Chinese Journal of Traumatology. 20(1): 1-3. https://doi.org/10.1016/j.cjtee.2016.05.004.
Hillger, F., Herr, G., Rudolph, R., and Schwarz, E. 2005. Biophysical comparison of BMP-2, proBMP-2, and the free pro-peptide reveals stabilization of the pro-peptide by the mature growth factor. Journal of Biological Chemistry. 280(15): 14974-14980. https://doi.org/10.1074/jbc.M414413200
Hogan, B.L. 1996. Bone morphogenetic proteins: Multifunctional regulators of vertebrate development. Genes & Development. 10(13): 1580-1594. https://doi.org/ 10.1101/gad.10.13.1580
Hyup Lee, J., Jang, S., Koo, T., Woo Suh, C., Lee, E.N., Lee, K., Lee, H.S., and Baek, H. 2011. Expression, purification and osteogenic bioactivity of recombinant human BMP-2 derived by Escherichia coli. Tissue Engineering and Regenerative Medicine. 8(1): 8-15.
Ishikawa, H., Kitoh, H., Sugiura, F., and Ishiguro, N. 2007. The effect of recombinant human bone morphogenetic protein-2 on the osteogenic potential of rat mesenchymal stem cells after several passages. Acta Orthop. 78(2): 285-292. https://doi.org/doi:10.1080/17453670710013816
Israel, D.I., Nove, J., Kerns, K.M., Moutsatsos, I.K., and Kaufman, R.J. 1992. Expression and characterization of bone morphogenetic protein-2 in chinese hamster ovary cells. Growth Factors. 7(2): 139-150. https://doi.org/10.3109/08977199209046403
Jayapal, K.P., Wlaschin, K.F., Yap, M.G.S., and Hu, W.S. 2007. Recombinant protein therapeutics from CHO cells-20 years and counting. Chemical Engineering Progress. 103:40-47.
Kimura, Y., Miyazaki, N., Hayashi, N., Otsuru, S., Tamai, K., Kaneda, Y., and Tabata, Y. 2010. Controlled release of bone morphogenetic protein-2 enhances recruitment of osteogenic progenitor cells for de novo generation of bone tissue. Tissue Engineering. 16(4): 1263-1270. https://doi.org/ 10.1089/ten.TEA.2009.0322
Kirker-Head, C.A. 2000. Potential applications and delivery strategies for bone morphogenetic proteins. Advanced Drug Delivery Reviews. 43(1): 65-92. https://doi.org/10.1016/S0169-409X(00)00078-8
Kirkham, G., and Cartmell, S. 2007. Genes and proteins involved in the regulation of osteogenesis. Ashammakhi N, Reis R, Chiellini E, editors. Topics in Tissue Engineering. 3.
Kubler, N.R., Reuther, J.F., Faller, G., Kirchner, T., Ruppert, R., and Sebald, W. 1998. Inductive properties of recombinant human BMP-2 produced in a bacterial expression system. International Journal of Oral and Maxillofacial Surgery. 27(4): 305-309. https://doi.org/10.1016/S0901-5027(05)80621-6
Lind, M., Eriksen, E.F., and Bünger, C. 1996. Bone morphogenetic protein-2 but not bone morphogenetic protein-4 and -6 stimulates chemotactic migration of human osteoblasts, human marrow osteoblasts, and u2-os cells. Bone. 18(1):53-57. https://doi.org10.1016/8756-3282(95)00423-8
Livak, K.J., and Schmittgen, T.D. 2001. Analysis of relative gene expression data using real-time quantitative pcr and the 2(-delta delta c(t)) method. Methods. 25(4):402-408. https://doi.org/doi:10.1006/meth.2001.1262
Lobstein, J., Emrich, C.A., Jeans, C., Faulkner, M., Riggs, P., and Berkmen, M. 2012. Shuffle, a novel escherichia coli protein expression strain capable of correctly folding disulfide bonded proteins in its cytoplasm. Microbial Cell Factories. 11:56. https://doi.org/doi:10.1186/1475-2859-11-56
Long, S., Truong, L., Bennett, K., Phillips, A., Wong-Staal, F., and Ma, H. 2006. Expression, purification, and renaturation of bone morphogenetic protein-2 from escherichia coli. Protein Expression and Purification. 46(2): 374-378. https://doi.org/ 10.1016/j.pep.2005.09.025
Luo, T., Zhang, W., Shi, B., Cheng, X., and Zhang, Y. 2012. Enhanced bone regeneration around dental implant with bone morphogenetic protein 2 gene and vascular endothelial growth factor protein delivery. Clinical Oral Implants Research. 23(4): 467-473. https://doi.org/10.1111/j.1600-0501.2011.02164.x
Marques, L.A.R.V., Júnior, E.A.C., Lotif, M.A.L., Neto, E.M.R., Silva, F.F.C., and Martiniano, C.R.Q. 2015. Application of BMP-2 for bone graft in dentistry. RSBO. 12(1): 88-93. https://doi.org/ 10.21726/rsbo.v12i1.176
Maruoka, Y., Oida, S., Iimura, T., Takeda, K., Asahina, I., Enomoto, S., and Sasaki, S. 1995. Production of functional human bone morphogenetic protein-2 using a baculovirus/Sf-9 insect cell system. Biochemistry and Molecular Biology International. 35(5): 957-963.
Martínez-Alonso, M., García-Fruitós, E., Ferrer-Miralles, N., Rinas, U., and Villaverde, A. 2010. Side effects of chaperone gene co-expression in recombinant protein production. Microbial Cell Factories. 9: 64. https://doi.org/ 10.1186/1475-2859-9-64
Massague, J. 1996. TGFbeta signaling: Receptors, transducers, and mad proteins. Cell. 85(7): 947-950. https://doi.org/10.1016/s0092-8674(00)81296-9
McKay, W.F., Peckham, S.M., and Badura, J.M. 2007. A comprehensive clinical review of recombinant human bone morphogenetic protein-2 (infuse bone graft). International Orthopaedics. 31(6): 729-734. https://doi.org/10.1007/s00264-0070418-6
Nasrabadi, D., Rezaeiani, S., Sayadmanesh, A., Baghaban, Eslaminejad M., and Shabani, A. 2018. Inclusion body expression and refolding of recombinant bone morphogenetic protein-2. Avicenna Journal of Medical Biotechnology. 10(4): 202-207.
Nissinen, L., Pirila, L., Heino, J. 1997. Bone morphogenetic protein-2 is a regulator of cell adhesion. Experimental Cell Research. 230(2): 377-385. https://doi.org/ 10.1006/excr.1996.3438
Poon, B., Kha, T., Tran, S., and Dass, C.R. 2016. Bone morphogenetic protein-2 and bone therapy: successes and pitfalls. Journal of Pharmacy Pharmacology. 68(2): 139-47. https://doi.org/10.1111/jphp.12506.
Pramesti, H.T., Suciati, T., Indrayati, A., Asjarie, S., and Retnoninggrum, D.S. 2012. Optimization of overproduction, solubilization, renaturation and its characterization. Biotechnology. 11(3): 133-143.
Quaasa, B., Burmeister, L., Li, Z., Nimtz, M., Hoffmann, A., and Rinas, U. 2018. Properties of dimeric, disulfide-linked rhBMP-2 recovered from E. coli derived inclusion bodies by mild extraction or chaotropic solubilization and subsequent refolding. Process Biochemistry. 67: 80-87. https://doi.org/10.1016/j.procbio.2018.02.001
Rane, A.M., Jonnalagadda, S., and Li, Z. 2013. On-column refolding of bone morphogenetic protein-2 using cation exchange resin. Protein Expression and Purification. 90(2): 135-140. https://doi.org/10.1016/j.pep.2013.05.008
Reddi, A.H. 2000. Morphogenesis and tissue engineering of bone and cartilage: Inductive signals, stem cells, and biomimetic biomaterials. Tissue Engineering. 6(4): 351-359. https://doi.org/10.1089/107632700418074
Retnoningrum, D.S., Pramesti, H.T., Santika, P.Y., Valerius, O., Asjarie, S., and Suciati, T. 2012. Codon optimization for high level expression of human bone morphogenetic protein-2 in Escherichia coli. Protein Expression and Purification. 84(2): 188-194. https://doi.org/10.1016/j.pep.2012.05.010
Ribeiro, M.M., Xu1, X., Klein, D., Kenyon, N.S., Ricordi, C., Felipe, M.S.S., and Pastori, R.L. 2010. Endotoxin deactivation by transient acidification. Cell Transplant. 19(8): 1047–1054. https://doi.org/10.3727/096368910X500643
Rosano, G.L., and Ceccarelli, E.A. 2014. Recombinant protein expression in Escherichia coli: advances and challenges. Front Microbiol, 5, 172. https://doi.org/10.3389/fmicb.2014.00172
Ruppert, R., Hoffmann, E., and Sebald, W. 1996. Human bone morphogenetic protein 2 contains a heparin-binding site which modifies its biological activity. European Journal of Biochemistry. 237(1): 295-302. https://doi.org/10.1111/j.14321033.1996.0295n.x
Scheufler, C., Sebald, W., and Hulsmeyer, M. 1999. Crystal structure of human bone morphogenetic protein-2 at 2.7 a resolution. Journal of Molecular Biology. 287(1): 103-115. https://doi.org/10.1006/jmbi.1999.2590
Sellers, R.S., Zhang, R., Glasson, S.S., Kim, H.D., Peluso, D., D'Augusta, D.A., Beckwith, K., and Morris, E.A. 2000. Repair of articular cartilage defects one year after treatment with recombinant human bone morphogenetic protein-2 (rhBMP-2). Journal of Bone and Joint Surgery. American. 82(2): 151-160.
Sharapova, N.E., Kotnova, A.P., Galushkina, Z.M., Lavrova, N.V., Poletaeva, N.N., Tukhvatulin, A.E., Semikhin, A.S., Gromov, A.V., Soboleva, L.A., and Ershova, A.S. et al. 2010. Production of the recombinant human bone morphogenetic protein-2 in Escherichia coli and testing of its biological activity in vitro and in vivo. Molecular Biology. 44(6): 923-930. https://doi.org/10.1134/s0026893310060099
Shimono, K., Oshima, M., Arakawa, H., Kimur, A., Nawachi, K., and Kuboki, T. 2010. The effect of growth factors for bone augmentation to enable dental implant placement: A systematic review. Japanese Dental Science Review. 46:43-53. https://doi.org/10.1016/j.jdsr.2009.10.004
Tang, P., Yao, Q., Zhang, W., Liang, Y., Zhang, L., and Wang, Y. 2009. A study of femoral neck fracture repair using a recombinant human bone morphogenetic protein-2 directional release system. Tissue Engineering Part A. 15(12): 3971-3978. https://doi.org/10.1089/ten.tea.2009.0052
Tuan, R.S., Boland, G., and Tuli, R. 2003. Adult mesenchymal stem cells and cell-based tissue engineering. Arthritis Research & Therapy. 5(1): 32-45.
Vallejo, L.F., Brokelmann, M., Marten, S., Trappe, S., Cabrera-Crespo, J., Hoffmann, A., Gross, G., Weich, H.A., and Rinas, U. 2002. Renaturation and purification of bone morphogenetic protein-2 produced as inclusion bodies in high-cell-density cultures of recombinant Escherichia coli. Journal of Biotechnoogy. 94(2): 185-194. https://doi.org/10.1016/S0168-1656(01)00425-4
Vaudel, M., Barsnes, H., Berven, F.S., Sickmann, A., and Martens, L. 2011. Searchgui: An open-source graphical user interface for simultaneous omssa and x!Tandem searches. Proteomics. 11(5): 996-999. https://doi.org/10.1002/pmic.201000595
Vaudel, M., Burkhart, J.M., Zahedi, R.P., Oveland, E., Berven, F.S., Sickmann, A., Martens, L., and Barsnes, H. 2015. Peptideshaker enables reanalysis of ms-derived proteomics data sets. Nature Biotechnology. 33(1): 22-24. https://doi.org/10.1038/nbt.3109
von Einem, S., Schwarz, E., and Rudolph, R. 2010. A novel two-step renaturation procedure for efficient production of recombinant bmp-2. Protein Expression and Purification. 73(1): 65-69. https://doi.org/10.1016/j.pep.2010.03.009
Wanachewin, O., Klangjorhor, J., Pothacharoen, P., Phitak, T., Laohapoonrungsee, A., Pruksakorn, D., and Kongtawelert, P. 2015. The promoting effects of sesamin on osteoblast differentiation of human mesenchymal stem cells. Journal of Functional Foods. 14: 395-406. https://doi.org/10.1016/j.jff.2015.01.046
Wang, E.A., Rosen, V., D'Alessandro, J.S., Bauduy, M., Cordes, P., Harada, T., Israel, D.I., Hewick, R.M., Kerns, K.M., and LaPan, P. et al. 1990. Recombinant human bone morphogenetic protein induces bone formation. Proceedings of the National Academy of Sciences of the United States of America. 87(6): 2220-2224. https://doi.org/10.1073/pnas.87.6.2220
Xiao, Y.T., Xiang, L.X., and Shao, J.Z. 2007. Bone morphogenetic protein. Biochemistry and Biophysical Research Communications. 362(3): 550-553. https://doi.org/10.1016/j.bbrc.2007.08.045
Zhang, Y., Ma, Y., Yang, M., Min, S., Yao, J., and Zhu, L. 2011. Expression, purification, and refolding of a recombinant human bone morphogenetic protein 2 in vitro. Protein Expression and Purification. 75(2): 155-160. https://doi.org/10.1016/j.pep.2010.07.014
Waraporn Kasekarn1*, Benjawan Suksiriphattanapong1, Tawan Chokepaichitkool2, Orawan Wanachewin3, Sittiruk Roytrakul4 and Prachya Kongtawelert2
1Department of Biochemistry, Faculty of Medical Science, Naresuan University, Phitsanulok 65000, Thailand
2Thailand Excellence Center for Tissue Engineering and Stem Cells, Department of Biochemistry, Faculty of Medicine, Chiang Mai University, Chiang Mai 50200, Thailand
3Program in Biology, Faculty of Science and Technology, Chiang Rai Rajabhat University, Chiang Rai 57100, Thailand
4Proteomics Research Laboratory, National Center for Genetic Engineering and Biotechnology, National Science and Technology Development Agency, Pathumthani 12120, Thailand
*Corresponding author. E-mail:wkasekarn@gmail.com, warapornkas@nu.ac.th
Total Article Views
Article history:
Received: September 2, 2019;
Revised: November 28, 2019;
Accepted: December 17, 2019

