
Identification of Single Nucleotide Polymorphism Markers Associated with Northern Corn Leaf Blight Resistance in Sweet Corn
Kedsadaporn Junta, Orapin Saritnum*, Ruangchai Juwattanasamran, Prawit Puddhanon, Darush Struss, Jumrearn Puttarach, and Seksan SongchanthuekPublished Date : 2019-12-09
DOI : https://doi.org/10.12982/CMUJNS.2020.0010
Journal Issues : Number 1, January-March 2020
ABSTRACT Northern corn leaf blight disease (NCLB) is a foliar disease of corn (Zea mays L.) caused by Exserohilum turcicum (Pass.). The Ht1, Ht2 and HtN1 genes in corn were found to control NCLB resistant traits. There has been an ongoing effort to identify various types of molecular markers associated with these genes in order to use them in marker-assisted selections. The objective of this study was to identify Single Nucleotide Polymorphism (SNP) markers which are associated with the Ht1 resistant gene. Ninety-three SNP markers were found in Ht1 regions, which are located on chromosome 2. Five SNP primers (MZSNP-0055106, MZSNP-0065744, MZSNP-0070164, MZSNP-0063922, MZSNP-0073150) showed polymorphism between susceptible and resistant lines. The Chi-square test of genotypic data of 184 F2 plants (NT58WS6#4 x ChallengerS6-1) amplified by five markers fit a 1:2:1 ratio with Chi-square values of 0.82, 1.08, 1.08, 0.64 and 0.64 respectively. These 5 SNP primers may be useful as molecular markers for NCLB resistance in sweet corn.
Keywords Northern corn leaf blight, Sweet corn, SNP markers, Resistance
INTRODUCTION
Northern corn leaf blight disease (NCLB), previously called Helminthosporium turcicum (Pass.), is a foliar disease of corn (Zea mays L.) which is caused by Exserohilum turcicum (Pass.) (Leonard et al., 1989). The most economically important host of E. turcicum is corn, especially in susceptible corn varieties. It could result in a yield loss of up to 30%. Optimal conditions for the disease infection include relatively low temperatures, such as 18-27 °C at night, and a humidity level of about 90-100% which is conducive to the growth of fungus (Ogliari et al., 2005). The disease symptoms appear as narrow lesions on leaves, expand to an elliptical shape and finally cause blight. Furthermore, symptoms may be found on the husks or leaf sheaths of susceptible corn hybrids. On the other hand, these lesions tend to be smaller due to reduced spore formation in resistant cultivars. In highly resistant hybrids, the only visible disease symptoms may be yellow spots.
Resistant cultivars have been widely used to control NCLB. E28 contains the gene Ht1, which confers resistance to E. turcicum, and was derived from a cross of Lv-9kuan with A619Ht1, followed by three backcrosses with Lv-9kuan and by gradual selection through self-pollination (Wu et al., 1996). There have been great efforts to explore the disease resistance of widely used maize lines in China, and 87.3-94.4% of these lines were found to be moderately sensitive or highly sensitive to this disease (Gao et al., 1997; Zhao, 2000). The HtNB dominant gene on chromosome 8 confers a non-lesion resistance at the flowering stage in an Indonesian landrace called ‘Bramadi’. HtNB has an independent hereditary pattern similar to Ht1, Ht2, and Ht3, and has a dominant epistatic effect, which establishes that the non-lesion resistance could be inhibiting the appearance of the chlorotic-lesion phenotype (Xu et al., 1987).
The resistant cultivars were used to provide an effective way to control NCLB. The identification of markers linked to resistance genes for sweet corn germplasm will facilitate the breeding of resistant sweet corn in Thailand. Currently, many researchers use molecular markers to assist with selection. One of the Thai sweet corn MAS studies, conducted by Puttarach et al. (2016), studied marker-assisted selections for resistance to NCLB. One highly susceptible line and three highly resistant lines were identified and used for population development. One hundred and fifty-seven F2 plants (NT58WS6#4 x ChallengerS6-1) were used to determine the linkage between traits and markers. Only two, SSR bnlg 1721 and umc1042 primers about 6.7 cM on chromosome 2, were closely linked to the resistant gene Ht1. Based on this result, these two SSR primers may be useful as molecular markers for NCLB resistance in sweet corn.
With the influence of PCR technology, primers that flank microsatellite loci are simple and fast to use, but the development of correctly functioning primers is an often tedious and pricey process. However, the Single Nucleotide Polymorphism (SNP) is a DNA sequence variation that occurs when a single nucleotide (A, T,C or G) differs among members of a species. SNPs are the most abundant marker system, both in animal and plant genomes, and have recently developed as the new generation molecular marker for several applications. Because of SNPs’ binary or co-dominant status, they are able to efficiently distinguish between homozygous and heterozygous alleles. Most significantly, SNPs are amenable to high throughput automation, allowing for a rapid and effective genotyping of large numbers of samples. Therefore, the objective of this study was to identify SNP markers associated with a Ht1 resistance gene in sweet corn. The SNP marker will be useful for marker-assisted selection in sweet corn breeding programs.
MATERIALS AND METHODS
Plant materials and DNA extraction
ChallengerS6-1 was the resistance line used in this study and the susceptible line was NT58WS6#4. F1 plants were derived from a cross between NT58WS6#4 x ChallengerS6-1 (S x R). The DNA was extracted from young leaves using a modified CTAB method (Nishiguchi et al., 2002).
SNP markers survey
SSR markers, either linked or located close to the known Ht1 gene found in the previous study (bnlg 1721 and umc 1042) (Puttarach et al., 2016), are detailed in Table 1. These two SSR markers were converted by a BLAST tool on the Maize Genetics and Genomics Database (2016) for data mining to SNP primers. Parental lines and the F1 population were screened with SNPs primer to examine polymorphic SNP markers. Finally, individual samples in the F2 population were genotyped by using polymorphic SNP markers that revealed differences between resistant and susceptible lines.
Table 1. SSR primers closely linked to NCLB resistance gene.
|
No. of Primers |
Gene |
Chromosome |
Gene Position |
|
12 |
Ht1 |
2 |
206,643,048 to 207,194,203 |
|
10 |
Ht2 |
8 |
135,858,136 to 398,042,717 |
|
10 |
HtN1 |
8 |
161,901,417 to 162,533,647 |
SNP genotyping assay
Each set of 100 µl SNP primers contained dH2O 46 µl, Forward primer 1 (F1) 12 µl, Forward primer 2 (F2) 12 µl and Reverse primer (R) 30 µl. A 1.6 µl PCR reaction contained DNA (15 ng/µl) 0.8 µl, KASPar master mix (2X ) 0.8 µl and SNP primer (54 µM) 0.02 µl. The amplification of DNA templates by PCR reaction involved predenaturing at 94 oC for 15 minutes, followed by touch down PCR with denaturing at 94 oC for 20 seconds, annealing at 65-57 oC for 10 rounds at 1 minute, PCR denaturing at 94 oC for 20 seconds and 36 rounds of annealing at 57 oC for 1 minute. PCR amplification included the following parameters. First, a competitive allelic specific primer FRET based assay called KASParTM genotyping system was used for SNP genotyping. Then, a fluorescence dye, VIC or FAM, was conjugated to primer and used in two FRET cassettes. A DNA sample was amplified with a thermal cycle using allele specific primers, leading to digression of the fluorescence dye and the quencher when the FRET cassette primer was hybridized to the DNA (Raitio et al., 2012).
RESULTS
SNP markers survey
Ninety-three SNP primers co-located with the known Ht1 genes converted from two SSR markers (bnlg 1721 and umc 1042) in the previous study (Puttarach et al., 2016) (Table 1) were screened for parental lines and F1 population. Only 5 SNP markers showed the polymorphic between parental lines and F1 samples. The scatter plot patterns of polymorphism of MZSNP-0055106, MZSNP-0065744, MZSNP-0070164, MZSNP-0063922 and MZSNP-0073150 among individuals of F2 population screening are shown in Figure 1A-1E.
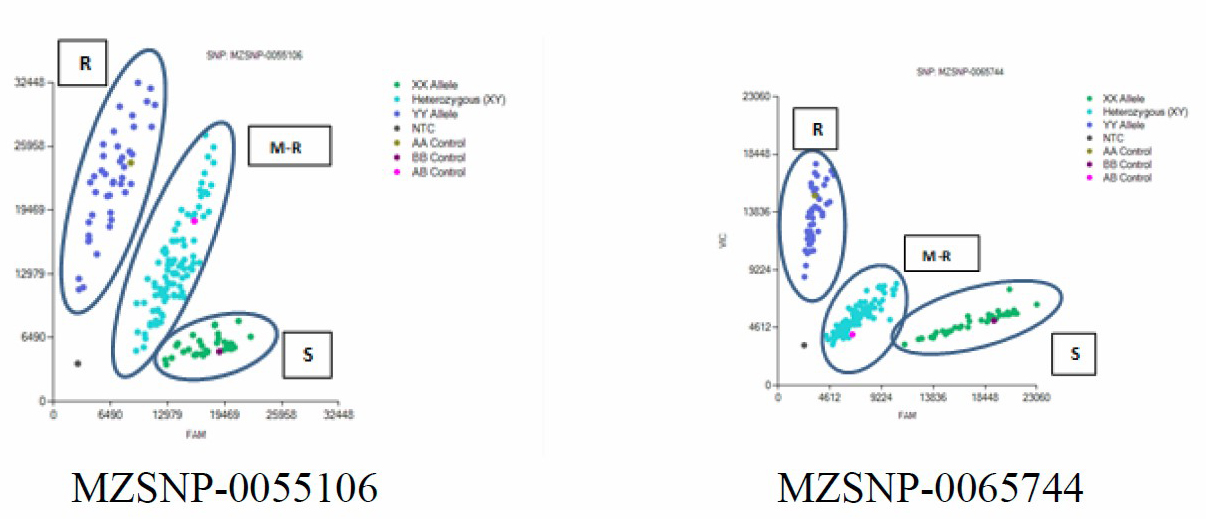
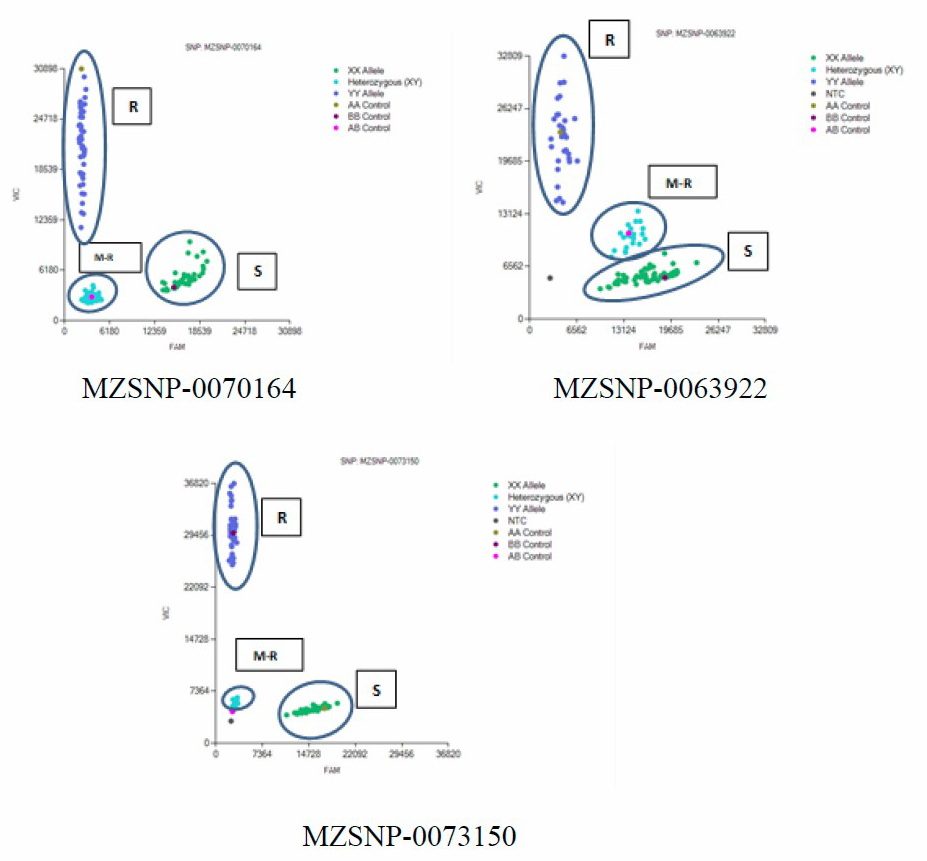
Figure 1. Scatter plot patterns. R = Resistance; M-R = Moderate resistance; S = Susceptible. A-E showed scatter plot chart of individuals of F2 population screening for MZSNP-0055106 (A), MZSNP-0065744 (B), MZSNP-0070164 (C), MZSNP-0063922 (D), MZSNP-0073150 (E) SNP primers.
Genotypic Chi-square analysis
The individual DNAs of an F2 population were distributed to homozygous susceptible parents (P1), homozygous resistant parents (P2) and the heterozygous (P1 x P2). The distinct genotypic classes were tested using a Chi-square analysis. The Chi-square values obtained were 0.82, 1.08, 1.08, 0.64 and 0.64 respectively. They were less than λ2 (P≤0.05) of 5.99 in the F2 population and fit in a 1:2:1 ratio (Table 2)
Table 2. Genotypic data of 184 individuals’ DNAs of F2 Population.

DISCUSSION
The ninety-three primers were deliberately chosen from Maize Genetics and Genomics Database (2016) by using a BLAST tool converting 2 SSR markers (bnlg 1721 and umc 1042) to a specific Ht1 gene. Yang (2010) reported that the flanking markers umc1042 and bnlg198 that were linked to a Ht1 gene in cultivar Ent17 were similar to the umc1042 that was closely linked to the Ht1 gene in ChallengerS6-1 (Puttarach et al.,2016). Moreover, Hooker (1963); Walz and Geiger (2000) reported that the Ht1 gene showed qualitative resistance and the gene action was partial dominance. These 5 SNP markers were closely linked with the Ht1 resistance gene (Figure 2) and the position of these SNP primers were also closely linked with the SSR markers (Table 3). The Chi-square test of the genotypic data of 184 F2 plants amplified by five markers was non-significantly or qualitatively fit in a 1:2:1 ratio (Table 1). Furthermore, the Chi-square values of genotypic data of these 2 SSR markers was also less than λ2 (P≤0.05) of 5.99 or non-significant (Puttarach et al., 2016). Therefore, the results of this study corresponded to the previous study. The results showed that these SNP markers could be used to assist in the selection of sweet corn germplasm possessing the resistance to NCLB disease in public and private sectors. Furthermore, these markers may be applied for gene pyramiding in Thai sweet corn breeding program for NCLB resistance.
Table 3. SNP primers closely linked to the NCLB resistance gene.
|
Marker |
Chr. |
Position |
P-value |
|
MZSNP-0055106 |
2 |
211,259,925 |
<0.0001** |
|
MZSNP-0065744 |
2 |
211,365,715 |
<0.0001** |
|
MZSNP-0070164 |
2 |
210,854,025 |
<0.0001** |
|
MZSNP-0063922 |
2 |
211,808,294 |
<0.0001** |
|
MZSNP-0073150 |
2 |
212,630,519 |
<0.0001** |
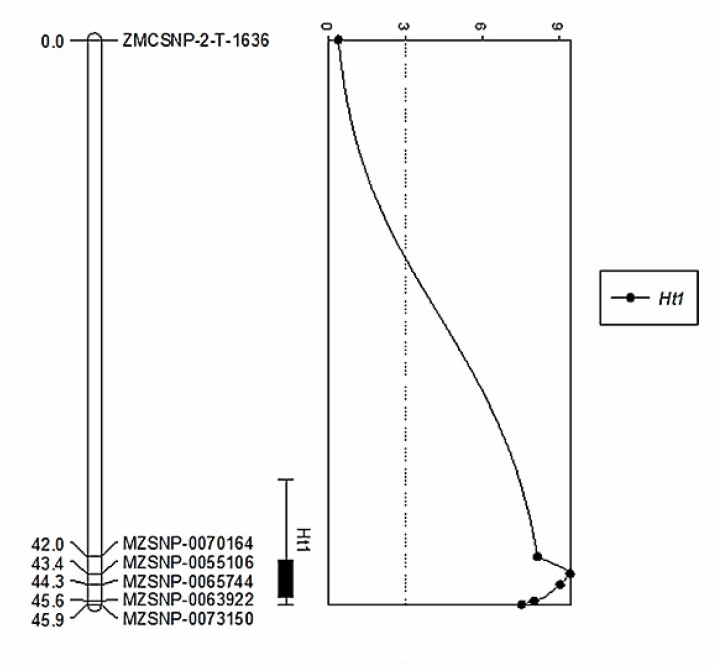
Figure 2. Genetic map for QTL controlling NCLB resistance (Ht1 gene).
CONCLUSION
The study was able to demonstrate that a Chi-square test of the genotypic data of 184 F2 population non-significantly or qualitatively fit in a 1:2:1 ratio. The Chi-square values of 0.82, 1.08, 1.08, 0.64 and 0.64, respectively were less than a λ2 (P≤0.05) of 5.99. This was consistent with NCLB resistance in sweet corn being conferred by one dominant gene.
ACKNOWLEDGEMENTS
This research is partially supported by the Center of Excellence on Agricultural Biotechnology, Science and Technology Postgraduate Education and Research Development office, office of Higher Education Commission, Ministry of Education (AG-BIO/PERDO-CHE). We also would like to thank Agronomy, Faculty of Agricultural Production, the office of Agricultural Research and Extension in Maejo University and the East West Seed Company for their cooperation and friendship.
REFERENCES
Gao, W.D., Dai, F.C., and Zhu X.Y. 1997. Evaluation of corn germplasm resources for resistance to four disease. Acta Phytotaxonom. Sinica. 24: 191-192.
Hooker, A.L. 1963. Inheritance of chlorotic-lesion resistance to Helminthosporium turcicum in seeding corn. Phytopathology. 53(6): 660-662.
Leonard, K.J., Levy, Y., and Smith D.R., 1989. Proposed nomenclature for pathogenic races of Exserohilum turcicum on corn. Plant Disease. 73: 776-777.
Nishiguchi, M.K., Doukakis, P., Egan, M., Kizirian, D., Phillips, A., Prendini, L., Rosenbaum, HC., Torres, E., Wyner, Y., DeSalle, R., et al. 2002. DNA isolation procedures. In: DeSalle, R., Giribet, G., Wheeler, W., editors. Techniques in molecular systematics and evolution. Switzerland: Birkhäuser Basel. p. 249-287.
Ogliari, J.B., Guimarães, M.A., Geraldi, I.O., and Camargo, L.E.A. 2005. New resistance genes in the Zea mays-Exserohilum turcicum pathosystem. Genetics and Molecular Biology. 28(3): 435-439
Puttarach, J., Puddhanon, P., Siripin, S., Sangtong, V., and Songchantuek, S. 2016. Marker assisted selection for resistance to northern corn leaf blight in sweet corn. SABRAO Journal of Breeding & Genetics. 48(1): 72-79.
Raitio, M., Perala, A., Harinen, R.R., and Lampinen, J. 2012. Validation of KASParTM dual FRET based SNP genotyping assay. Finland: Thermo Fisher Scientific. p. 1-4.
Welz, H.G., and Geiger, H.H. 2000. Genes for resistance to northern corn leaf blight in diverse maize populations. Plant Breeding. 119(1): 1-14.
Wu, J.C., Chen, G., Zhang, T.Y., and Liu, C.Z. 1996. Application of multiple resistance inbred line E28 of corn in breeding. Journal of Maize Scinces. 4: 1-4.
Xu, S.Z., Liu, S.D., Xiong, X.Z., and Liu, J.L. 1987. Indonesian landrace ‘Bramadi’selected lines, the resistant heredity mechanism of northern corn leaf blight. Scientia Agricultura Sinica. 20: 48-55.
Yang, E. 2010. Dissection of resistant genes to northern corn leaf blight in CIMMYT maize cultivar Entl7 and bioinformatics analysis of candidate sequences between two flanking markers linked with resistance gene Htl. Journal of Agricultural Science.
Zhao, B.R. 2000. Evaluation of corn germplasm resources for resistance to northern corn leaf blight. Journal of Maize Scinces. 8: 91-92.
Kedsadaporn Junta1, 5, Orapin Saritnum2, 5*, Ruangchai Juwattanasamran1, Prawit Puddhanon1, 5, Darush Struss3, Jumrearn Puttarach5, and Seksan Songchanthuek4
1Department of Agronomy, Faculty of Agricultural Production, Maejo University, Chiang Mai 50290, Thailand
2Department of Horticulture, Faculty of Agricultural Production, Maejo University, Chiang Mai 50290, Thailand
3Department of Biotechnology, East West Seed Company, Chiang Mai 50290, Thailand
4The office of Agricultural Research and Extension, Maejo University, Chiang Mai 50290, Thailand
5Center of Excellence on Agricultural Biotechnology: (AG-BIO/PERDO-CHE), Bangkok 10900, Thailand
*Corresponding author. E-mail: orapins343@hotmail.com
Total Article Views
Article history:
Received: October 9, 2018;
Revised: May 28, 2019;
Accepted: June 11, 2019

