
First Report of Chromosome Analysis of Two Siganid Fishes (Perciformes, Siganidae)
Kriengkrai Seetapan and Sitthisak Pinmongkhonkul*Published Date : 2019-12-09
DOI : https://doi.org/10.12982/CMUJNS.2020.0001
Journal Issues : Number 1, January-March 2020
ABSTRACT This is the first report of chromosome analysis in the white- spotted rabbitfish(Siganus canalicutus) and the orange-spotted spinefoot (S. gattatus) from the Andaman Sea, Thailand. Kidney cell samples were taken from three male and three female fishes. The mitotic chromosome preparation was created through direct engineering on kidney cells. Conventional and Ag-NOR staining techniques were applied so as to stain the chromosomes. The results showed that the diploid chromosome number of both S. canalicutus and S. gattatus was 2n=48, that the fundamental number (NF) was 50 in S. canalicutus and 54 in S. gattatus. Among the chromosomes present, both acrocentric and telocentric ones were identified as 2- 46 and 6- 42 respectively. No heteromorphic chromosomes were observed which could be considered as sex-chromosomes. After Ag- NORs banding technique, a single pair of nucleolar organiser regions/ NORs was observed on the short arm telomeric region of large acrocentric chromosome Pair 1 in both species. The karyotype formulas could be deduced as:
S. canalicutus 2n (48) = La2+Lt22+Mt16+St8
S. gattatus 2n (48) = La4+Lt18+Ma2+Mt22+St2
Keywords: Siganus canalicutus, Siganus gattatus, Karyotype, Chromosome
INTRODUCTION
The family Siganidae (Rabbitfishes, Spinefoot) is classified into the order Perciformes. The genus Siganus is the only one member of the family. It comprises 21 species, namely Signanus argenteus, S. corallinus, S. doliatus, S. canaliculatus, S. fuscescens, S. guttatus, S. javus, S. labyrinthodes, S. lineatus, S. niger, S. puellus, S. punctatus, S. punctatissimus, S. randalli, S. spinus, S. stellatus, S. unimaculatus, S. uspi, S. vergatus, S. vermiculatus, and S. vulpinus (Allen et al., 1999). Fishes in the family Siganidae are generally characterised by a laterally compressed body which is shaped like a torpedo, a small jaw and a dorsal fin with thirteen strong spines and ten soft rays. This fin is also preceded by a short, sharp, forwardly projecting spine, usually protruding slightly from its pocket but sometimes completely hidden. They have an anal fin with seven strong spines and nine rays. The pelvic fins comprise seven spines, a character unique to this family. All dorsal, anal and pelvic fin spines are grooved and contain venom glands. If handled carelessly, they are capable of inflicting very painful wounds. They have very small cycloid scales, sometimes absent from isthmus and midthoracic regions, but if present on the head, they don't extend beyond the suborbital area of the cheek. As for their colour, most species are usually brightly colored and ornately patterned whereas other ones are often drab and tend to become mottled with brown at death (Carpenter and Niem, 2001).
There were approximately eight percent of the Perciformes order that were recorded for their karyological characteristics, revealing a model with a diploid chromosome number of 2n= 48 ( Affonso et al., 2001) . However, different karyotypes from the typical order Perciformes pattern had frequently been detected, indicating Robertsonian rearrangements as the preferential process in some groups, such as the families Labridae and Pomacentridae (Ueno and Takai, 2000; Molina and Galetti, 2002). The cytogenetical reports on the family Siganidae demonstrated a highly conserved pattern, considered basal for the order Perciformes ( 2n= 48) , except for S. spinus ( 2n= 42) ( Kitada et al., 1979; Choudhury et al., 1979; Ojima and Yamamoto, 1990; Jumrusthanasan et al., 2015). No study describing the karyotypes of the white- spotted rabbitfish ( Siganus canalicutus) and orange- spotted spinefoot ( S. gattatus) had been published previously. Almost, these fishes are importance species as food and sometime, they are kept in aquarium as ornamental fish but the knowledge of these species has no data especially biology and cytogenetics. These present study is the first report of cytogenetic studies of S. canalicutus and S. gattatus performed using conventional staining and Ag- NOR banding techniques. The obtained results attempt to provide further cytogenetic information for future studies on taxonomy and evolutionary relationships between these fishes.
MATERIAL AND METHODS
Materials
Three males and three females S. canalicutus and S. gattatus were obtained from the Andaman Sea in Southern Thailand (Figure 1). These fishes were transferred to some laboratory aquaria and were kept under standard conditions for three days prior to the experiment. Isolation procedures involving fish chromosomes were carried out directly on kidney cells (Supiwong et al., 2012; 2013; 2015). The chromosome preparations were stained with 10% Giemsa for 30 minutes and NORs were identified by Ag-NOR staining (Howell and Black, 1980). The metaphase figures were analysed according to the chromosome classification after Turpin and Lejeune (1965). The centromeric index (CI) for chromosomes situated in the ranges 0.50-0.59, 0.60-0.69, 0.70-0.89, and 0.90-0.99 were described as metacentric, submetacentric, acrocentric, and telocentric respectively. The Fundamental number (NF) was obtained by assigning a value of two to metacentric, submetacentric and acrocentric chromosomes and a value of one to telocentric chromosomes. Thirty metaphase plates per specimen were used for classify chromosome types.
The classify for chromosome sizes is as:
L > mean of LT of the largest pair + mean of LT of the smallest pair
2
M ≤ mean of LT of the largest pair + mean of LT of the smallest pair
2
S < mean of LT of the largest pair
2
RESULTS
Diploid number, fundamental number and karyotype of S. canalicutus and S. gattatus
The present cytogenetic description on type and size of chromosome was done by using the conventional staining and Ag-NOR banding techniques. The findings show that S. canalicutus and S. guttatus have the modal 2n of 48 and the respective NF of 50 and 54 in both sexes (Figure 2). There was no observation of unusual size chromosome related to sex for all analysed species. This report constitutes the first mention of the modal of 2n=48 for the pertinent members of the family Siganidae.
The karyotype formulas for S. canalicutus and S. gattatus are as follows:
S. canalicutus 2n (48) = La2+Lt22+Mt16+St8
S. gattatus 2n (48) = La4+Lt18+Ma2+Mt22+St2
However, there are no significant differences in chromosome size between ‘large’ and ‘medium’ and between ‘medium’ and ‘small’, the presentation in Table 2 and 3 is not recommended.
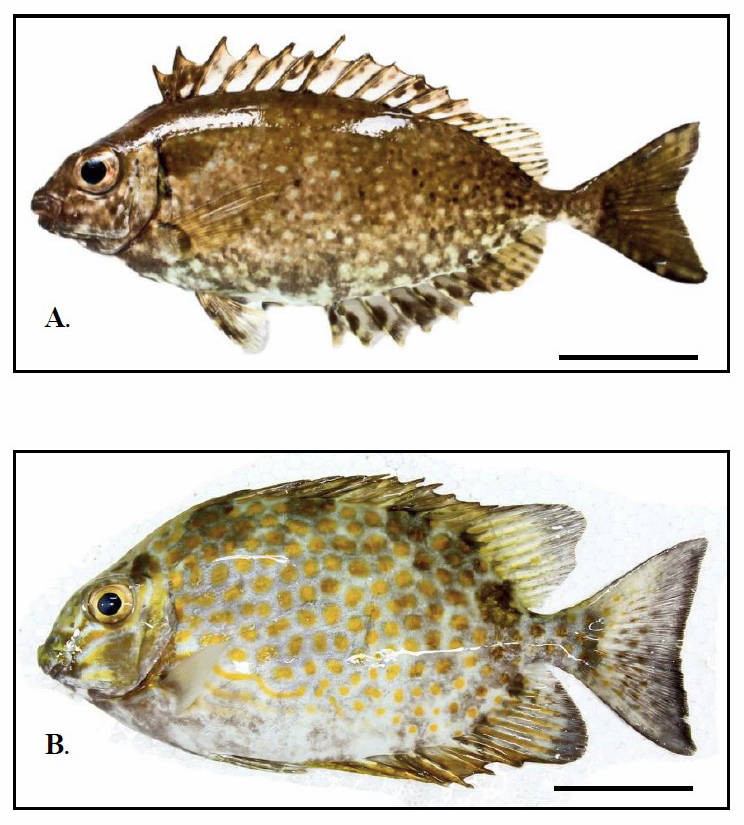
Figure 1. General characteristics of Siganus canalicutus (A.) and S. gattatus (B.). Scale bars indicate five centimeters.
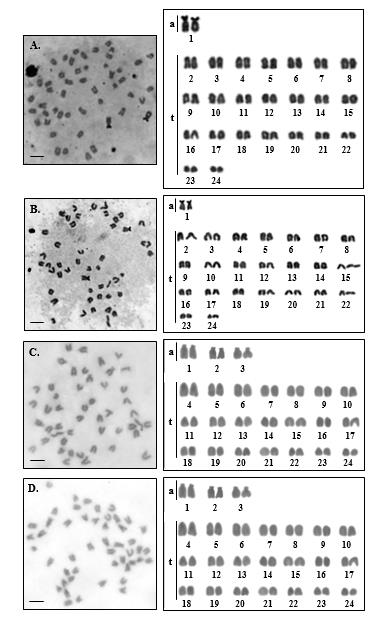
Figure 2. Metaphase chromosome plates and karyotypes of male ( A. ) and female ( B. ) Siganus canalicutus, 2n= 48 and male ( C. ) and female ( D.) S. gattatus, 2n= 48 by conventional straining technique. Scale bars indicate five micrometers.
Chromosome markers of S. canalicutus and S. gattatus
The objective of Ag- NOR banding technique is to reach out the nucleolar organizer region (NOR) which is the representative location of genes (loci) that function in ribosome synthesis ( 18S and 28S ribosomal RNA) ( Sharma et al. , 2002; Khakhong et al., 2014). If these are regions are active during the interphase prior to mitosis, they could be detected by silver nitrate staining ( Howell and Black, 1980) since it specifically stains a set of acidic protein related to ribosomal synthesis process; this technique actually reveals active NORs ( Ag- NORs) and not the rDNA associated to NORs (Jordan, 1987).
Through Ag- NORs banding technique, a single pair of NOR was observed on the short arm telomeric region of the large acrocentric chromosome pair 1 in both species ( Figures 3). The chromosome markers determination of these two species showed the positive Ag-NOR marks on pairs of homologous chromosomes. They are distinguished into the group depending on NOR locations which have the short arm of telomeric NOR.
The asymmetrical karyotypes of S. canalicutus and S. gattatus, and the only two types of chromosomes ( acrocentric and telocentric) that were found, are important chromosome markers. The idiogram shows continuous length gradation of chromosomes. The largest and smallest chromosomes present size differences (approximately threefolds). Data of chromosomal checks on mitotic metaphase cells of S. canalicutus and S. gattatus are shown in Tables 1, 2 and 3, respectively. Figures 4 and 5 show the idiograms obtained by conventional staining and Ag-NOR banding techniques, respectively.
According to previous reports, there are four species in the family Siganidae, namely S. fuscescens, S. javus, S. spinus and S. vermiculatus which were analysed for their karyotype information. The present study, the hypothesis model for the chromosome evolution of the family Siganidae on the basis of cytogenetic descriptions of six fishes including S. canalicutus, S. fuscescens, S. gattatus, S. javus, S. spinus and S. vermiculatus is proposed (Figures 6).
Table 1. Review of siganid fishes cytogenetic reports (Perciformes, Siganidae).
|
Species |
2n |
NF |
Karyotype |
NORs |
Locality |
Reference |
|
Siganus canalicutus S. fuscescens S. gattatus S. javus S. spinus S. vermiculatus |
48 48 48 48 42 48 |
50 50 54 48 48 50 |
2a+46t 2a+46t 6a+42t 48t 6m+36t 2a+46t |
1(TR) N/A 1(TR) N/A N/A 1(TR) |
Thailand Japan Thailand Japan Japan Thailand |
Present study Kitada et al. (1979) Present study Choudhury et al. (1979) Ojima and Yamamoto (1990) Jumrusthanasan et al. (2015) |
Notes: 2n = diploid chromosome number, NF = fundamental number ( number of chromosome arms) , m = metacentric chromosome, a = acrocentric chromosome, t = telocentric chromosome, NORs = nucleolar organizer regions, TR = telomeric region, and N/A = not analysed yet.
Table 2. Mean length of short arm chromosome ( Ls) , long arm chromosome (Ll), total arm chromosome (LT), relative length (RL), and centromeric index (CI) from 20 metaphases of the white-spotted rabbitfish (Siganus canaliculatus), 2n=48.
|
Chro. pair |
Ls |
Ll |
LT |
RL |
CI |
Chro. Size |
Chro. type |
|
1* |
0.23 |
0.61 |
0.84 |
0.065 |
0.724 |
Large |
Acrocentric |
|
2 |
0.00 |
0.72 |
0.72 |
0.056 |
1.000 |
Large |
Telocentric |
|
3 |
0.00 |
0.69 |
0.69 |
0.053 |
1.000 |
Large |
Telocentric |
|
4 |
0.00 |
0.67 |
0.67 |
0.052 |
1.000 |
Large |
Telocentric |
|
5 |
0.00 |
0.65 |
0.65 |
0.050 |
1.000 |
Large |
Telocentric |
|
6 |
0.00 |
0.64 |
0.64 |
0.049 |
1.000 |
Large |
Telocentric |
|
7 |
0.00 |
0.62 |
0.62 |
0.048 |
1.000 |
Large |
Telocentric |
|
8 |
0.00 |
0.60 |
0.60 |
0.04 |
1.000 |
Large |
Telocentric |
|
9 |
0.00 |
0.58 |
0.58 |
0.045 |
1.000 |
Large |
Telocentric |
|
10 |
0.00 |
0.56 |
0.56 |
0.043 |
1.000 |
Large |
Telocentric |
|
11 |
0.00 |
0.55 |
0.55 |
0.042 |
1.000 |
Large |
Telocentric |
|
12 |
0.00 |
0.55 |
0.55 |
0.042 |
1.000 |
Large |
Telocentric |
|
13 |
0.00 |
0.54 |
0.54 |
0.042 |
1.000 |
Medium |
Telocentric |
|
14 |
0.00 |
0.53 |
0.53 |
0.041 |
1.000 |
Medium |
Telocentric |
|
15 |
0.00 |
0.52 |
0.52 |
0.040 |
1.000 |
Medium |
Telocentric |
|
16 |
0.00 |
0.51 |
0.51 |
0.039 |
1.000 |
Medium |
Telocentric |
|
17 |
0.00 |
0.49 |
0.49 |
0.038 |
1.000 |
Medium |
Telocentric |
|
18 |
0.00 |
0.48 |
0.48 |
0.037 |
1.000 |
Medium |
Telocentric |
|
19 |
0.00 |
0.47 |
0.47 |
0.036 |
1.000 |
Medium |
Telocentric |
|
20 |
0.00 |
0.42 |
0.42 |
0.032 |
1.000 |
Medium |
Telocentric |
|
21 |
0.00 |
0.40 |
0.40 |
0.031 |
1.000 |
Small |
Telocentric |
|
22 |
0.00 |
0.36 |
0.36 |
0.028 |
1.000 |
Small |
Telocentric |
|
23 |
0.00 |
0.33 |
0.33 |
0.025 |
1.000 |
Small |
Telocentric |
|
24 |
0.00 |
0.26 |
0.26 |
0.020 |
1.000 |
Small |
Telocentric |
Notes: * = NOR-bearing chromosome (satellite chromosome) and chro. = chromosome.
Table 3. Mean length of short arm chromosome (Ls), long arm chromosome (Ll), total arm chromosome ( LT) , relative length ( RL) , and centromeric index (CI) from 20 metaphases of the orange-spotted spinefoot (Siganus guttatus), 2n=48.
|
Chro. pair |
Ls |
Ll |
LT |
RL |
CI |
Chro. Size |
Chro. type |
|
1* |
0.270 |
0.708 |
0.978 |
0.055 |
0.724 |
Large |
Acrocentric |
|
2 |
0.216 |
0.624 |
0.840 |
0.048 |
0.743 |
Large |
Acrocentric |
|
3 |
0.195 |
0.518 |
0.713 |
0.04 |
0.727 |
Medium |
Acrocentric |
|
4 |
0.000 |
0.875 |
0.875 |
0.05 |
1.000 |
Large |
Telocentric |
|
5 |
0.000 |
0.850 |
0.850 |
0.048 |
1.000 |
Large |
Telocentric |
|
6 |
0.000 |
0.838 |
0.838 |
0.047 |
1.000 |
Large |
Telocentric |
|
7 |
0.000 |
0.818 |
0.818 |
0.046 |
1.000 |
Large |
Telocentric |
|
8 |
0.000 |
0.798 |
0.798 |
0.045 |
1.000 |
Large |
Telocentric |
|
9 |
0.000 |
0.775 |
0.775 |
0.044 |
1.000 |
Large |
Telocentric |
|
10 |
0.000 |
0.765 |
0.765 |
0.043 |
1.000 |
Large |
Telocentric |
|
11 |
0.000 |
0.763 |
0.763 |
0.043 |
1.000 |
Large |
Telocentric |
|
12 |
0.000 |
0.733 |
0.733 |
0.041 |
1.000 |
Large |
Telocentric |
|
13 |
0.000 |
0.725 |
0.725 |
0.041 |
1.000 |
Medium |
Telocentric |
|
14 |
0.000 |
0.715 |
0.715 |
0.04 |
1.000 |
Medium |
Telocentric |
|
15 |
0.000 |
0.713 |
0.713 |
0.04 |
1.000 |
Medium |
Telocentric |
|
16 |
0.000 |
0.705 |
0.705 |
0.04 |
1.000 |
Medium |
Telocentric |
|
17 |
0.000 |
0.690 |
0.690 |
0.039 |
1.000 |
Medium |
Telocentric |
|
18 |
0.000 |
0.673 |
0.673 |
0.038 |
1.000 |
Medium |
Telocentric |
|
19 |
0.000 |
0.650 |
0.650 |
0.037 |
1.000 |
Medium |
Telocentric |
|
20 |
0.000 |
0.643 |
0.643 |
0.036 |
1.000 |
Medium |
Telocentric |
|
21 |
0.000 |
0.603 |
0.603 |
0.034 |
1.000 |
Medium |
Telocentric |
|
22 |
0.000 |
0.588 |
0.588 |
0.033 |
1.000 |
Medium |
Telocentric |
|
23 |
0.000 |
0.558 |
0.558 |
0.031 |
1.000 |
Medium |
Telocentric |
|
24 |
0.000 |
0.475 |
0.475 |
0.027 |
1.000 |
Small |
Telocentric |
Notes: * = NOR-bearing chromosome (satellite chromosome) and chro. = chromosome.
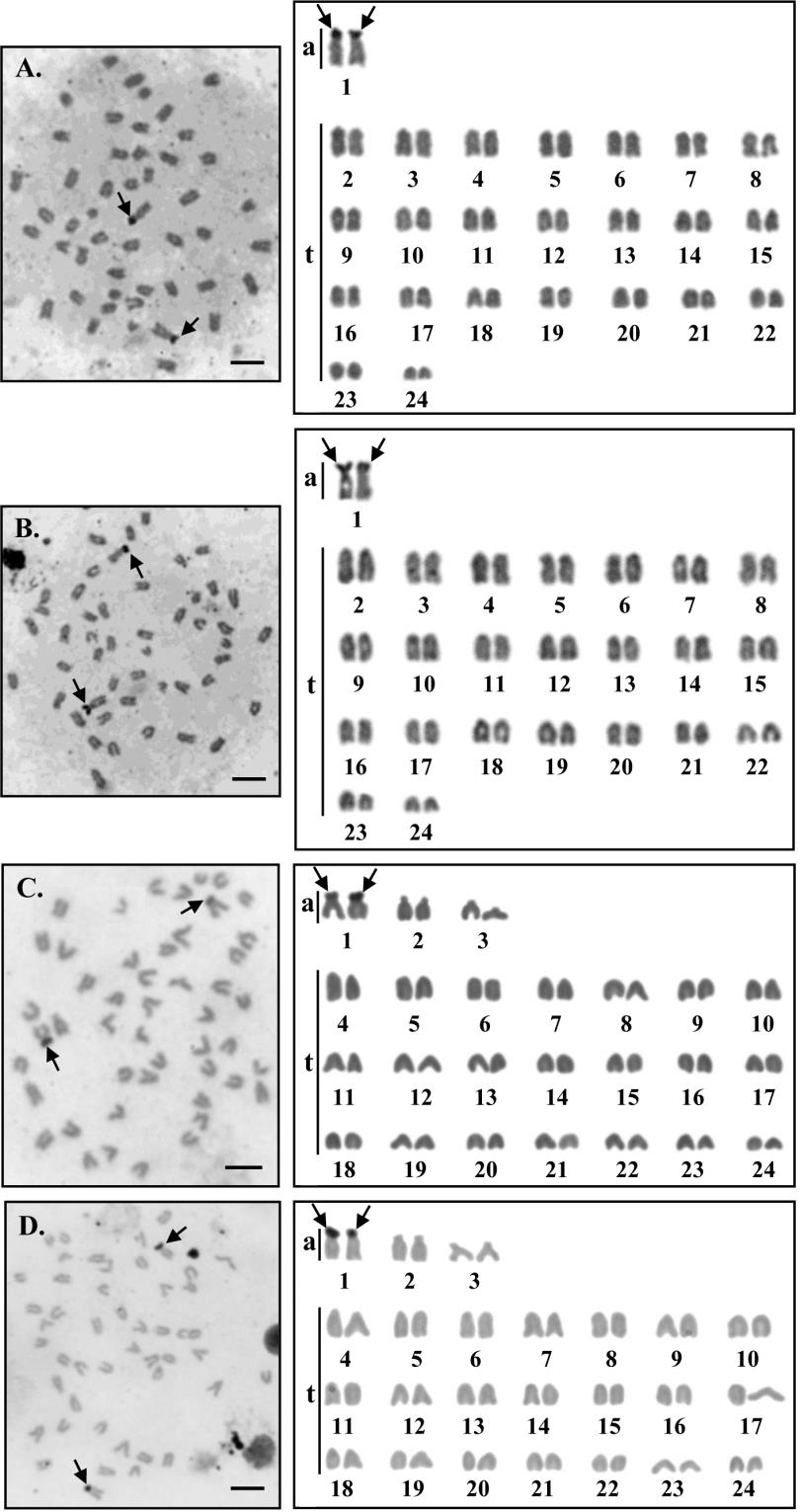
Figure 3. Metaphase chromosome plates and karyotypes of male (A.) and female ( B. ) Siganus canalicutus, 2n= 48 and male ( C. ) and female ( D. ) S. gattatus, 2n= 48 by Ag- NOR banding technique. Arrows indicate NORs bearing (scale bars = 5 micrometers).
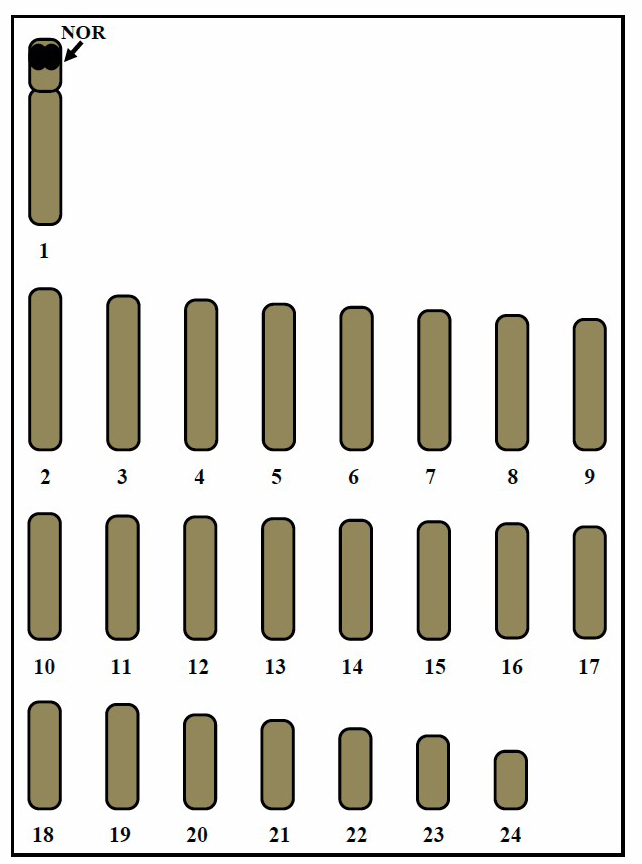
Figure 4. Idiogram showing lengths and shapes of chromosomes of Siganus canalicutus, 2n=48. Arrow indicates NORs bearing chromosome pair 1.
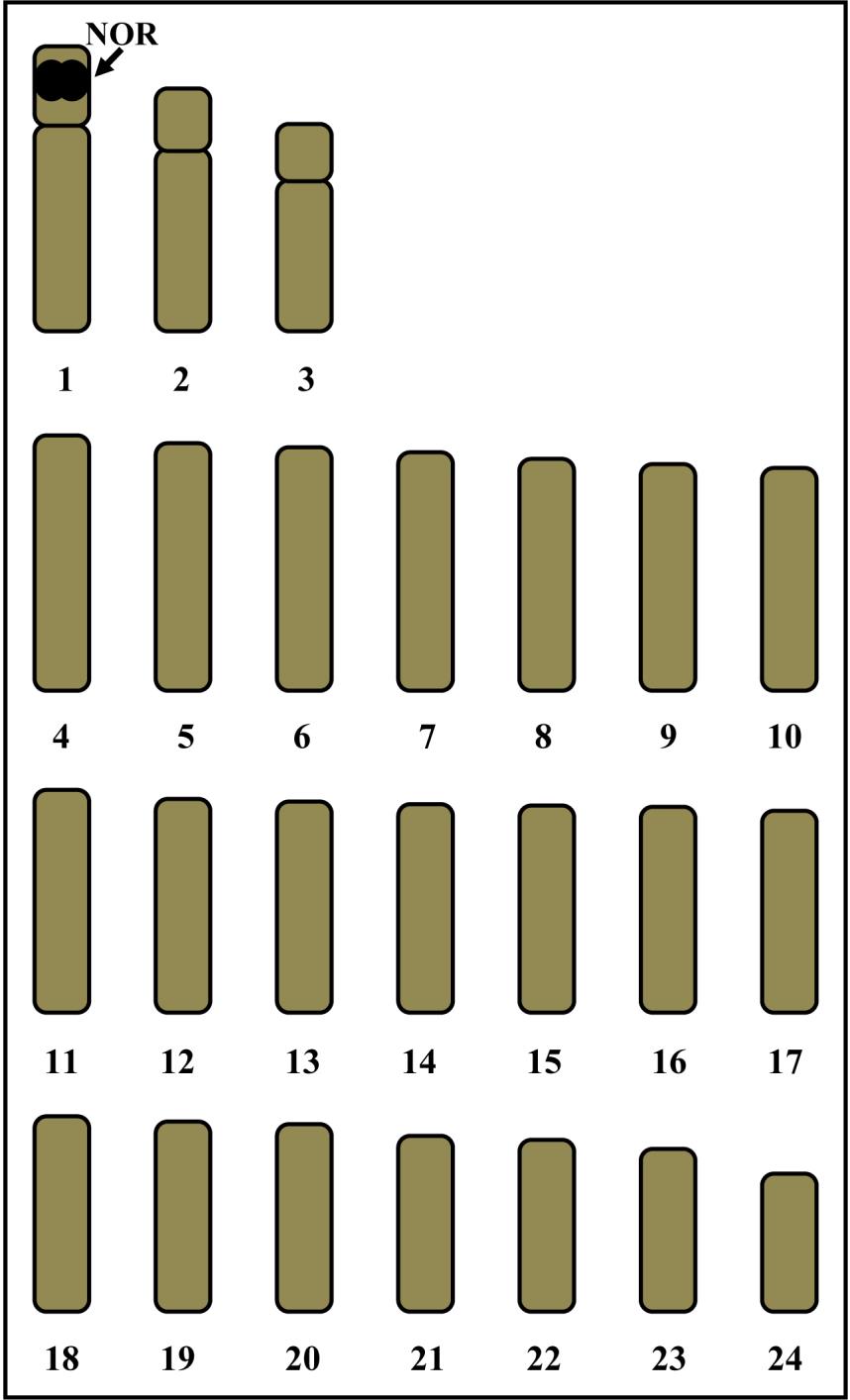
Figure 5. Idiogram showing lengths and shapes of chromosomes of Siganus gattatus, 2n=48. Arrow indicates NORs bearing chromosome pair 1.
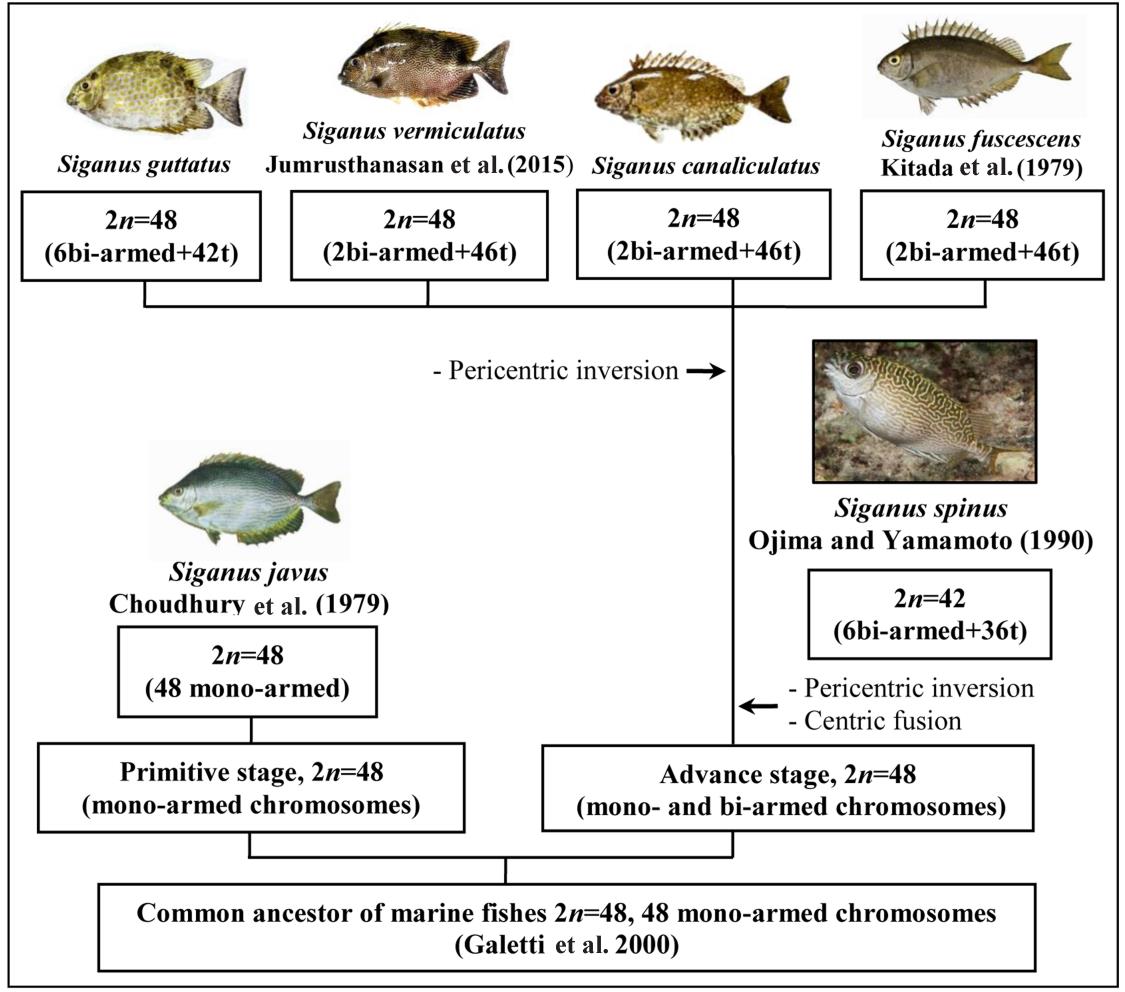
Figure 6. The hypothesis model for the chromosome evolution of the family Siganidae.
DISCUSSION
In the present study, the findings pertaining to S. canalicutus and S. guttatus are consistent with those published in the chromosomal characteristic reports on the Siganidae fishes including S. fuscescens ( Kitada et al., 1979) , S. javus (Choudhury et al., 1979) and S. vermiculatus (Jumrusthanasan et al., 2015). However, different information on the diploid chromosome number was found in a chromosomal examination of S. spinus which revealed the 2n of 42 (Ojima and Yamamoto, 1990). The NF was 50 in S. canalicutus and 54 in S. gattatus. The differences of karyological characteristics between both species show variations in the number of chromosome arms which are similar to those stated in the previous reports of NF for the other siganid fishes, namely S. fuscescens, S. javus, S. spinus, and S. vermiculatus that have their NF ranging from 48 to 50 (Choudhury et al., 1979; Kitada et al., 1979; Ojima and Yamamoto, 1990; Jumrusthanasan et al., 2015).
Both investigated species have no cytologically distinguishable sex chromosome. This characteristic is similar to other ones in the family Siganidae (Choudhury et al., 1979; Kitada et al., 1979; Ojima and Yamamoto, 1990; Jumrusthanasan et al., 2015). It is possible that the fish’s sex- chromosomes are dependent upon an initiation of differentiation. Therefore, chromosomes containing sex- determination genes cannot be found by cytogenetic analyses (Bertollo et al., 2004; Kaewmad et al., 2014; Kasiroek et al., 2017).
Chromosome markers of S. canalicutus and S. gattatus, the location of NOR in a terminal position and close to the centromeres are also founded upon a primitive feature ( Vitturi et al., 1988) . NOR is frequently used to compare variations, as well as to identify and explain specifications. Species, which have limited gene exchange due to geographical isolation, have elevated karyotype numbers and NOR variations. The use of NORs in explaining kinships depends on a large extent on the uniformity of this characteristic and on the degree of variety within a taxon (Yüksel and Gaffaroğlu, 2008).
The cytogenetic information which was obtained in this research, i.e. 2n, NF, type and sizes of chromosomes of two analysed fishes of the family Siganidae, namely S. canalicutus and S. gattatus exhibited all of them have the same diploid chromosomes number of 48. Nevertheless, differences in NF and type as well as size of chromosome were as shown. These variations on each member’s chromosomal characteristics revealed the occurrence of chromosomal evolution among the species. Although most Siganid species have an NF of 48 or 50, with most telocentric chromosomes in the complement, the number of chromosome arms in the Siganus genus varies from 48 to 54. Under the assumption that species with a larger NF are more advanced in evolutionary terms, the most recent species within the Siganus genus would be S. fuscescens, NF=50; S. javus, NF=48 and S. vermiculatus, NF=50. However, despite differences in NF, all Siganus species share the same diploid chromosome number (2n=48). Such changes in chromosome arm number appear to be related to the occurrence of pericentric inversions, which are among the most common modifications contributing to karyotypic rearrangement in fishes and other vertebrates (Galetti et al., 2000).
That is the good recommendation. However, we discuss the chromosomal evolution in this group, only species that have been cytogenetic reported. We only hypothesize the trend of chromosomal evolution which is possible to occur in this family if there are more data from more species, they will be good hypothesis for chromosomal evolution.
CONCLUSION
The diploid chromosome number of S. canalicutus and S. gattatus as 2n= 48. The chromosomes were identified as belonging to the acrocentric and telocentric types defined as 2- 46 and 6- 42, respectively. No heteromorphic chromosomes were observed that could be considered as sex- chromosomes. Following an Ag- NORs banding technique, a single pair of nucleolar organiser regions/ NORs was observed on the short arm telomeric region of large acrocentric chromosome pair 1 in both species. The karyotype formulas could be deduced as:
S. canalicutus 2n (48) = La2+Lt22+Mt16+St8
S. gattatus 2n (48) = La4+Lt18+Ma2+Mt22+St2
ACKNOWLEDGEMENTS
This research was supported by grants of Unit of Excellence 2019 on Biodiversity and Natural Resources Management, University of Phayao (UoE62005). The authors thank Professor Dr.Alongklod Tanomtong, Khon Kaen University for laboratory support.
REFERENCES
Affonso, P.R.A.M., Guedes, W., Pauls, E., and Galetti, P.M., Jr. 2001. Cytogenetic analysis of coral reefs fishes from Brazil (families Pomacanthidae and Chaetodontidae). Cytologia. 66(4): 379–384. https:// doi.org/10.1508/cytologia.66.379
Allen, G., Swainston, R., and Ruse, J. 1999. Marine fishes of South-East Asia. Singapore: Berkeley Books.
Bertollo, L.A.C., Oliveira, C., Molina, W.F., Margarido, V.P., Fontes, M.S.M., Pastori, C., Falcão, J.N., and Fenocchio, A.S. 2004. Chromosome evolution in the erythrinid fish, Erythrinus erythrinus (Teleostei: Characiformes). Heredity. 93: 228–233. https://doi.org/10.1038/sj.hdy.6800.511
Carpenter, K.E., and Niem V.H. 2001. The living marine resources of the Western Central Pacific. Rome: Food and Agriculture Organization of the United Nations.
Choudhury, R.C., Prasad, R., and Das, C.C. 1979. Chromosomes of six species of marine fishes. Caryologia. 32(1): 15–21. https://doi.org/10.1080/ 00087114.1979.10796773
Galetti, P.M.Jr., Aguilar, C.T., and Molina, W.F. 2000. An overview on marine fish cytogenetics. Hydrobiologia. 420: 55–62. https://doi.org/10.1023/ A:1003977418900
Howell, W.M., and Black, D.A. 1980. Controlled silverstaining of nucleolus organizer regions with a protective colloidal developer: a 1-step method. Experientia. 36(8): 1014–1015. https://doi.org/10.1007/BF01953855
Jordan, G. 1987. At the heart of the nucleolus. Nature. 329: 489–490. Jumrusthanasan, S., Supiwong, W., Suvarnaraksha, A., Jiwyam, W.,
Pinthong, K., and Tanomtong, A. 2015. First karyological analysis of the vermiculate spinefoot, Siganus vermiculatus (Perciformes, Siganidae) from Thailand. Cytologia. 80(1): 111–116. https://doi.org/10.1508/ cytologia.80.111
Kasiroek, W., Indananda, C., Luangoon, N., Pinthong, K., Supiwong, W., and Tanomtong, A. 2017. First chromosome analysis of the humpback cardinalfish, Fibramia lateralis (Perciformes, Apogonidae). Cytologia. 82(1) Special Issue: 9–15. https://doi.org/10.1508/cytologia.82.9
Kaewmad, P., Monthatong, M., Supiwong, W., Saowakoon, S., and Tanomtong, A. 2014. Natural autotetraploid and chromosomal characteristics in the subfamily botiinae (Cypriniformes, Cobitinae) from Northeast Thailand. Cytologia. 79(3): 299–313. https://doi.org/10.1508/ cytologia.79.299
Khakhong, S., Supiwong, W., Tanomtong, A., Sriuttha, M., Jearranaiprepame, P., Soemphol, W., and Jiwyam, W. 2014. A first chromosomal characterization of NORs in splendid snakehead fish, Channa lucius (Perciformes, Channidae). Cytologia. 79(2): 133–139. https://doi.org/ 10.1508/cytologia.79.133
Kitada, J., Tatewaki, R., and Tagawa, M. 1979. The karyotype of the common aigo, Siganus fuscescens. Chromosome Information Service. 27: 12–13.
Molina, W.F., and Galetti, P.M. 2002. Robertsonian rearrangement in the reef fish Chromis (Perciformes, Pomacentridae) involving chromosomes bearing 5S rRNA genes. Genetics and Molecular Biology. 25(4): 373– 377. https://doi.org/10.1590/S1415-4757200200040004
Ojima, Y., and Yamamoto, K. 1990. Cellular DNA contents of fishes determined by flow cytometry. La Kromosomo II. 57: 1871–1888.
Sharma, O.P., Tripathi, N.K., and Sharma, K.K. 2002. A review of chromosome banding in fishes. In: Sobti, R.C., Obe, G., and Athwal, R.S., editors. Some aspects of chromosome structure and functions. Delhi: New Narosa Publishing House.
Supiwong, W., Tanomtong, A., Khakhong, S. Silawong, K., Aoki, S., and Sanoamuang, L. 2012. The first chromosome characteristics of nucleolar organizer regions and karyological analysis of clown knife fish, Chitala ornata (Osteoglossiformes, Notopteridae) by T-lymphocyte cell culture. Cytologia. 77: 393–399. https://doi.org/10.1508/cytologia.77.393
Supiwong, W., Tanomtong, A., Jumrusthanasan, S., Khakhong, S., Neeratanaphan, L., and Sanoamuang, L. 2013. Standardized karyotype and idiogram of titan triggerfish, Balistoides viridescens (Tetraodontiformes, Balistidae) in Thailand. Cytologia. 78(4): 345-351. https://doi.org/10.1508/cytologia.78.345
Supiwong, W., Tanomtong, A., Pinthong, K., Kaewmad, P., Poungnak, P., and Jangsuwan, N. 2015. The first chromosomal characteristics of nucleolar organizer regions and karyological analysis of pink anemonefish, Amphiprion perideraion (Perciformes, Amphiprioninae). Cytologia. 80: 271–278. https://doi.org/10.1508/cytologia.80.271
Turpin, R., and Lejeune, J. 1965. Les chromosomes humains. Paris: Gauther Villars.
Ueno, K., and Takai, A. 2000. Chromosome evolution involving Robertsonian rearrangements in Xirichthys fish (Labridae, Perciformes). Cytobios. 103: 7–15.
Vitturi, R., Libertini, A., Mazzola, A., Colomba, M.S., and Sara, G. 1996. Characterization of mitotic chromosomes of four species of the genus Diplodus: karyotypes and chromosomal nucleolar organizer regions phenotypes. Journal of Fish Biology. 49(6): 1128–1137. https://doi.org/ 10.1111/j.1095-8649.1996.tb01783.x
Yüksel, E. and Gaffaroğlu, M. 2008. The analysis of nucleolar organizer regions in Chalcalburnus mossulensis (Pisces: Cyprinidae). Journal of Fisheries Science. 2: 587–291.
Kriengkrai Seetapan1 and Sitthisak Pinmongkhonkul2*
1School of Fishery, Faculty of Agriculture and Natural Resources, University of Phayao, Phayao 56000, Thailand
2School of Biology, Faculty of Science, University of Phayao, Phayao 56000, Thailand
*Corresponding author. E-mail: Sitthisak.pi@up.ac.th
Total Article Views
Article history:
Received: January 7, 2019;
Revised: April 18, 2019;
Accepted: May 1, 2019

