
Partial Sequence Analysis of Cytochrome b Gene by FINS Technique Reveals Fraud Sambar Meat in Wild Food Restaurant Thanat Vorajinda, Chavin Chaisongkram, Wibhu Kutanan, and Khemika Lomthaisong*
Published Date : 2019-09-20
DOI : https://doi.org/10.12982/CMUJNS.2019.0031
Journal Issues :
Number 4 , October-December 2019
ABSTRACT
Species identification of animals in the Cervidae by sequence compar- ison of the Cytochrome b (Cyt b) gene using FINS method is focused in this study. The Cyt b fragments of seven species in the Cervidae, including rusa deer (Cervus timorensis), sambar deer (C. unicolor), sika deer (C. nippon), hog deer (C. porcinus), axis deer (C. axis), Eld’s deer (C. eldi) and barking deer (Munticacus muntjak) were amplified by our own designed primers. The amplicons (322 bp) were sequenced and their partial sequences (263 bp) were then analyzed. Barking deer showed the highest value of genetic diversity within species (π = 0.0364; h = 0.9). The phylogenetic analysis had shown that the partial sequence of Cyt b gene can be used to classify most studied species in Cervidae accurately, except for sambar and rusa deer that cannot be differentiated. Hence, species identification of unknown meat samples was then performed by this method. Referenced Cyt b sequences of wild boar (Sus scrofa), favorite meat in wild food restaurant, and the Cyt b sequence of known sambar tissue samples were additionally compared. The genetic distances indicated that unknown meat samples were presumably wild boar. Although this method cannot differentiate sambar from rusa deer, this study will be useful for wildlife forensic particularly when screening examination of irrelevant samples and fraud sambar meats identification are necessary.
Keywords: Cytochrome b gene, DNA variation, Cervidae, FINS
INTRODUCTION
Nowadays, large numbers of endangered wildlife animals listed in the CITES appendices are smuggled for illegal trades. In this case, wildlife forensic science has been used as a significant tool to prosecute the smuggler. Studies on wildlife forensic science have been reviewed (Johnson et al., 2014). Most of them have focused on species identification, for which methods based on DNA analysis are more advantageous than those of traditional morphological identification because they can be applied on different types of samples, for example, processed animal parts, derivatives within Traditional Chinese Medicine (TCMs) and objects made from animal parts (Iyengar, 2014) DNA markers used for species identification must be species-specific. These sequences can be found in both nuclear and mitochondrial DNA. However, mitochondrial DNA is more preferable because it contains high copy numbers and hardly degrades.
Many studies have shown that species-specific sequences of mitochondrial DNA, such as ND1 (Kitpipit et al., 2012; Welton et al., 2013), COI (Wilson-Wilde et al., 2010), 16S rRNA (Imaizumi et al., 2007), Cyt b (Jun et al., 2011), D-loop (Fumagalli et al., 2009; Gupta et al., 2011) and ITS-2 (Clarke et al., 2006) can be used to identify animal species. Molecular techniques like PCR-RFLP have been conducted to discriminate DNA polymorphisms of a target sequence for species identification, for example Cyt b for identification of fish species in the Cyprinidae family (Chen et al., 2011) and 16S rRNA for sea cucumber identification (Wen et al., 2010). However, the questioned species may not be revealed when the PCR-RFLP profile of an unknown sample does not match with any of those reference species. Hence, a technique called FINS (Forensically Informative Nucleotide Sequencing) has been introduced. This technique identifies species by using phylogenetic analysis in which DNA sequence of a sample is compared with known DNA sequences in the database (Li et al., 2011)). The FINS technique has been reported as a successful method for determination of authentic seafood ingredients including, octopus (Espineira et al., 2010), jelly fish (Armani et al., 2013) and ling fish (Santaclara et al., 2014). This technique has also been applied for mislabeling investigation of pet canned food (Armani et al., 2015).
In wildlife forensic science, species identification based on DNA analysis can be performed by PCR amplification of target DNA using species-specific primer, i.e. examination of a shawl woven from Tibetan antelope hair, a protected species listed in the CITES appendix I (Lee et al., 2006) and identification of protected buffalo meat in Sri Lanka (Rajapaksha et al., 2003). Nevertheless, species identification by PCR amplification with species-specific primer cannot differentiate samples of different subspecies, in which case determination of genetic variation by FINS will cope with this limitation as shown by Gupta et al. (2013) who employed the technique to distinguish wild pigs from domestic ones. However, species identification by FINS needs reference DNA data from related species. The lack of reference DNA may cause misinterpretation. Unknown samples of a viverrid were misidentified as coming from hyena due to the insufficient data of Veverricula indica 16S rRNA sequences in the NCBI database (Sahajpal and Goyal, 2010). In Thailand, large numbers of livestock belong to the Cervidae family, i.e. sika, rusa, hog and sambar deer. The latter is a protected species; however, due to the popularity of its meat, farming sambar deer is allowed albeit only through legal permission. Since its farming practice is subject to tight regulation, the samba deer meat is highly valued and costly. Owing to this, meats of other animals are sometimes deceitfully advertised as the meat of sambar deer, necessitating proper identification of the meats. Although DNA barcoding (COI) has been successfully used to identify sambar deer meat, the accuracy of this method depends on the quality of the database (Kumar et al., 2012). Moreover, for species identification by DNA barcoding, a target size of 5’ COI region is approximately 650 bp (Trivedi et al., 2016). This may be troublesome when highly degraded DNA samples are performed. Hence, an alternative method for sambar deer meat identification by comparing Cyt b sequences using FINS technique is focused in this study. According to our results, the FINS technique has proved a useful tool for species identification enabling us to forensically trace the origins of the meats sold in wildlife restaurants across Thailand.
MATERIALS AND METHODS
Samples
Blood samples of five individuals per species including C. timorensis, C. unicolor, C. nippon, C. porcinus, C. axis, C. eldi and M. muntjak were kindly provided by Khon Kaen Zoo under the permission from the Animal Ethics Committee (ACUC-KKU-31/2559). Ten milliliters of blood was taken into a test tube containing 500 µl of 0.5M EDTA. Blood samples were then kept at 4°C until DNA extraction was performed. For unknown meats, five samples (100 g each) were taken from wild food restaurants in Thailand (two from Prajeenburi, two from Phra Nakorn Sri Ayuttaya and one from Nakhon Nayok province). The meats were kept at 4°C until they were used. For reference, meat (MC) and liver (LC) of C. unicolor and C. timorensis antler (HC) were also provided by Khon Kaen Zoo.
DNA isolation
DNA extraction from the blood samples was conducted using QIAamp® DNA Blood Mini Kit (QIAGEN, Germany). Briefly, 200 µl of each sample was incubated with proteinase K at 56°C for 10 min. Then, ethanol was added and the mixture was transferred onto QIAamp mini spin column. This column was centrifuged to get rid of the liquid. The column was subsequently washed twice. The DNA was then eluted from the column. For meat and antler samples, DNA extraction using NucleoSpin® kit was performed according to the manufacturer’s instruction. Twenty-five mg of each tissue sample was incubated with proteinase K at 56°C for 3 h. Then, 100% ethanol was added and the mixture mixed before being transferred to NucleoSpin® tissue column. The liquid was removed from the column by centrifugation. The column was then washed and DNA was subsequently eluted. The quantity and quality of DNA solution were examined by NanoDrop spectrophotometer.
Primer design
The nucleotide sequences of Cyt b genes from animals in the Cervidae were obtained from GenBank database (http://www.ncbi.nlm.nih.gov/genbank). Cyt b sequences of C. porcinus (DQ379301.1), C. axis (AY607040.1), C. eldi (AY157735.1), C. nippon (D32192.1), C. timorensis (AF423200.1), C. unicolor (AF423201.1) and M. muntijak (AF042718.1) were multiple aligned using ClustalW2 program (http://www.ebi.ac.uk/Tools/msa/clustalw2/). Primers were designed from conserved regions by OligoAnalyzer 3.1 (https://www.idtdna. com/calc/analyzer).
PCR amplification
The PCR reaction containing 0.2 ng DNA, 0.4 µM of each primer and 1xPCR Master Mix (Vivantis, Malysia) was set up. A total volume of 25 µl was made by adding deionized water. The amplification was then performed in a Thermal Cycler with the following cycling conditions: initial denaturation step at 94oC for 2 min followed by 35 cycles of 94oC 30 sec, 57oC 30 sec, 72oC 30 sec and a final extension at 72oC 5 min. The PCR product was subsequently analyzed on 1.5% agarose gel. The size of the amplicon was estimated from the 100 bp DNA marker (VIVANTIS, Malaysia).
DNA sequencing and haplotype analysis
The PCR products were sent to Macrogen (South Korea) for DNA sequencing with our designed primers. The nucleotide sequences of forward and reverse strands from each sample were aligned by BioEdit 7.0 program (Hall, 1999). Fine adjustment was manually performed with visual inspec- tion. These sequences were then assembled. The sequences of all samples were multiple aligned. Fault insertion and deletion of the bases were manually edited. Nucleotide sequences were trimmed at both ends to produce the equal lengths of the sequences. From the result by multiple alignment, the polymorphic sites of nucleotide were examined using DnaSP 5.0 (Librado and Rozas, 2009). Then, nucleotide diversity (π) and haplotype diversity (h) were determined.
Phylogenetic analysis
Thirty-five sequences of the Cyt b gene obtained from this study were aligned with seven sequences of species belonging to the Cervidae familyretrieved from the GenBank database, using MEGA version 6. A fragment of 263 bp Cyt b gene was selected for comparison by means of neighbor-joining (NJ) tree. The genetic distances were also computed using K2-parameter model with 2000 bootstrap re-samplings.
Species identification by FINS methods
The DNA extraction from unknown meat samples, reference samples of C. unicolor (meat, liver) and C. timorensis (antler) were conducted. PCR amplification of the Cyt b gene was then performed followed by DNA sequencing of amplicon as described earlier. The DNA sequences were compared with reference sequences of the Cyt b gene of known cervids.
RESULTS
Primer design
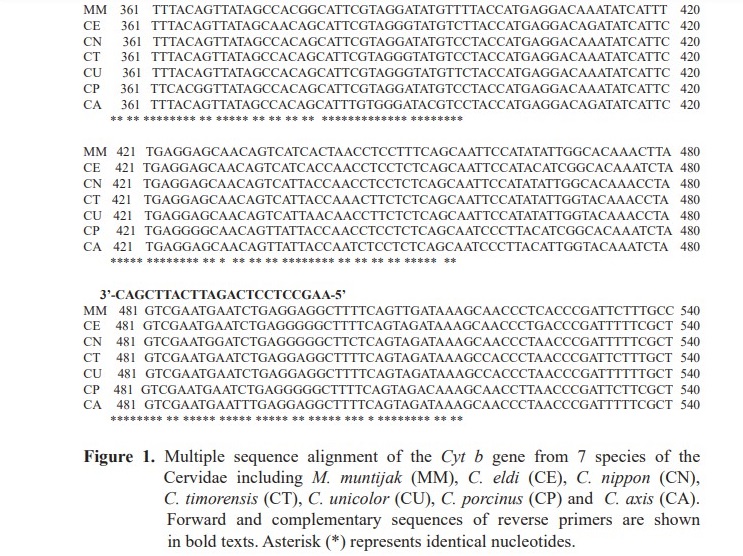
Based on the result from the alignment of Cyt b gene sequences from seven species of the Cervidae, primers were designed from conserved regions. Then, nucleotide positioned 182-200 and 481-503 were chosen for forward and reverse primers, respectively (Figure 1). As a result, a DNA fragment with a length of 322 bp was amplified.


PCR results
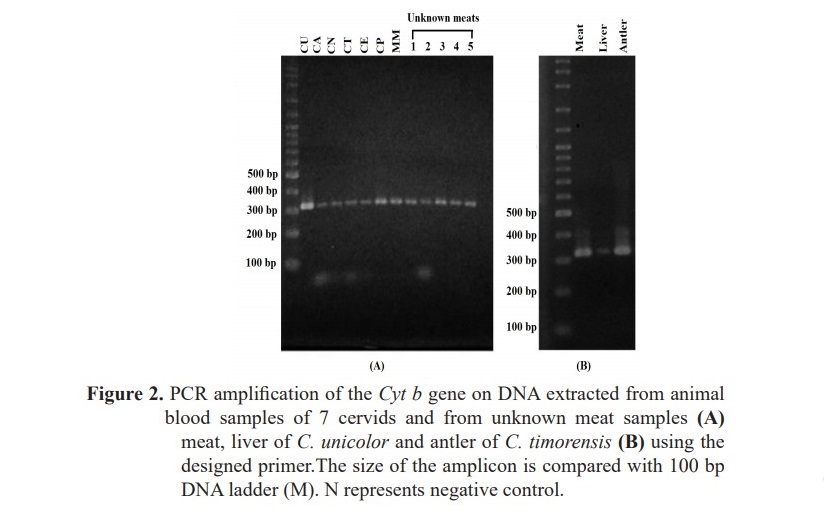
DNAs extracted from the animal blood and meat samples were quantified and diluted to 50 ng/ul for PCR amplification. Optimal melting temperature for PCR was also investigated. As expected from the primer design result, the amplicon was 322 bp long in all samples (Figure 2), indicating successful primer design.
Haplotype diversity
The 263 bp nucleotide sequences of 322 bp PCR products from each species were analyzed for haplotype and nucleotide diversity. Unexpectedly, the Cyt b gene from C. unicolor, C. timorensis, C. nippon and C. axis samples had no sequence variation as indicated by one haplotype in each species (Table 1). On the contrary, the DNA sequences of the Cyt b gene from M. muntjak samples showed highest variation (h = 0.9; π = 0.03640) in which four haplo- types were identified. For C. eldi and C. porcinus, two haplotypes were found.
Table 1. Number of haplotypes, haplotype diversity and nucleotide diversity in the Cyt b gene (263 bp) of the cervids.

Phylogenetic analysis
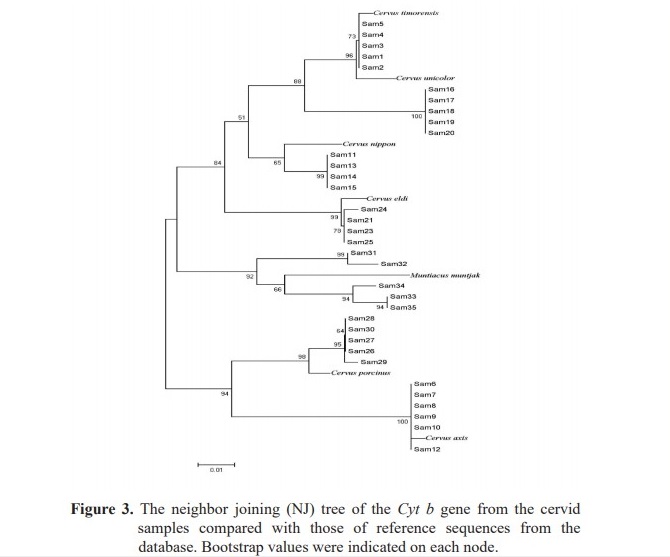
The 263 bp nucleotide sequences of the Cyt b gene from 35 samples were compared and analyzed whether or not they could be used for species identification. In order to prove this, Cyt b reference sequences of the cervids retrieved from the GenBank database were also included in the NJ tree (Figure 3). Noteworthy, samples of 5 out of 7 species were classified in the same group as their reference species indicating that the sequence of this Cyt b region could be used as DNA target for species identification. Although, the two species of C. unicolor and C. timorensis could not be differentiated, most species in the Cervidae could be identified. Moreover, sample no.12 was grouped as C. axis instead of C. nippon. Although, this sample was subjected to repeated analyses, the same result was achieved indicating the possibility of mislabeling.
Species identification
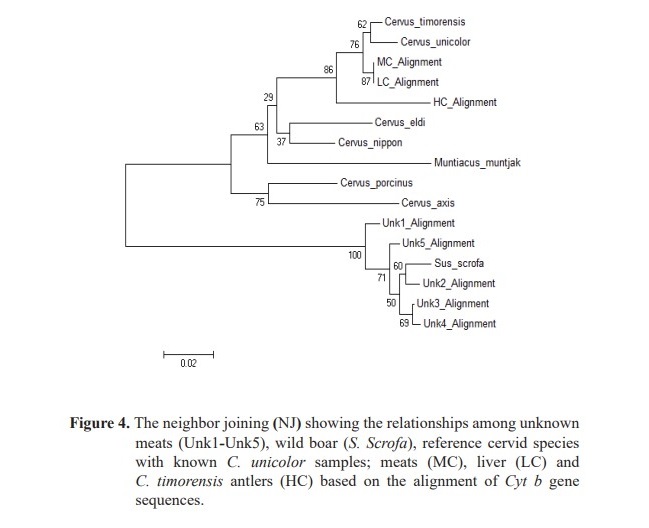
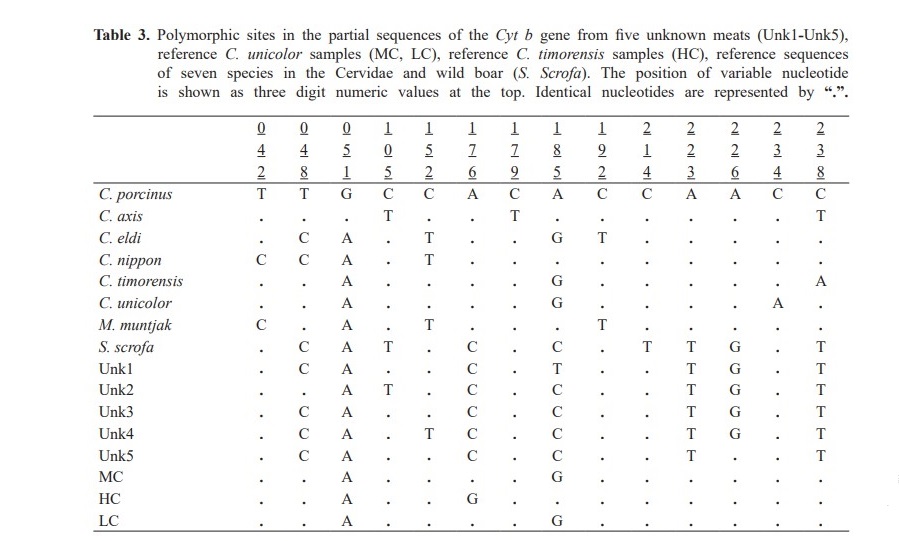
For FINS technique, the DNA sequences (263 bp) from unknown meats were compared with the reference sequences of Cyt b from seven species of the Cervidae and known C. unicolor samples. Additionally, wild boar (S. Scrofa), a popular meat ingredient in wild food restaurants was also included to represent a fraud sambar meat occurrence. Phylogenetic analysis revealed that unknown meats (Unk1-Unk5) were clustered in the same group as wild boar (Figure 4). The pairwise distances are shown in Table 2. The variable nucleo- tide positions of Cyt b sequences from sambar deer and wild boar are shown in Table 3.

DISCUSSION
Species identification of sambar deer (C. Unicolor), a member of the Cervidae which is listed in Thailand Wildlife Protection, by FINS technique had been examined in this study. A species-specific sequence of Cyt b gene was focused because it has been reported as the locus most frequently used for species identification in forensic science (Parson et al., 2000; Hsieh et al., 2001). Hence, a pair of primers were designed from the Cyt b sequences of seven species of the Cervidae. PCR amplification of Cyt b by these primers was performed on 35 DNA samples of the cervids (5 samples/species). These primers were appropriate to this study as an amplicon of Cyt b (322 bp) was produced in all samples (Figure 1). The sequence analysis of these amplicons was then conducted. Interestingly, the Cyt b sequences of M. muntjak showed highest DNA variations, followed by C. eldi and C. porcinus, respectively (Table 1). However, no DNA variations were observed among sequences of the rest of the species. Low genetic variation in this case may be explained by the inbreeding because they have been raised in an enclosed zoo. Studies on species identification by Cyt b sequence has been reported earlier but the larger amplified DNA fragments of 358 bp (Parson et al., 2000), 402 (Hsieh et al., 2003) and 421 (Gupta et al., 2013) were investigated.
The NJ tree was then constructed to analyze whether these Cyt b sequences could determine the species in the Cervidae (Figure 2). The partial sequences of Cyt b (263 bp) were compared with those of reference cervids. Noticeably,samples were classified in the same group as their source species except for samples from C. unicolor and C. timorensis that cannot be differentiated. These results indicated that the Cyt b sequences (263 bp) could be used as a marker for most species identification in the Cervidae. Hence, unknown meats from wild food restaurants were determined for their species using FINS analysis on these sequences (Figure 3). Surprisingly, we found that all unknown meats were classified in the same group as wild boar. To confirm the results that the unknown meats were not belong to Cervidae, controlled samples of C. unicolor samples were also investigated and they were clustered in the same group as C. unicolor and C. timorensis. Three forensically informative nucle- otide variations were observed between unknown meats and S. scrofa (Table 3). In addition, BLAST analysis was also performed in order to ensure that our investigations were correct. We found that the sequences of unknown meats (Unk1-5) were close to wild boar with similarity at 94%, 96%, 96%, 96% and 96%, respectively. These evidences clearly confirm that the analysis of Cyt b (263 bp) by the FINS method can identify the source species of meat.
This study has shown that comparison of Cyt b partial sequence (263 bp) by the FINS technique is useful for species identification not only for cervids but also other species (if reference sequences are available). Even though, the differentiation between C. unicolor and C. timorensis cannot be made, this method still has the advantage for wildlife forensic in which it can be used to exclude the irrelevant evidence from consideration. Moreover, as the shorter targeted Cyt b had been investigated in this study, DNA analysis for species identification on degraded samples could be achieved (Lopez-Oceja et al., 2016).
CONCLUSION
Comparison of Cyt b partial sequence (263 bp) by FINS method can be used for species identification of animals in the Cervidae. However, C. unicolor and C. timorensis cannot be discriminated by this method. For wildlife forensic application, this study may be useful for preliminary examination to exclude irrelevant evidence.
REFERENCES
Armani, A., Tinacci, L., Giusti, A., Castigliego, L., Gianfaldoni, D., and Guidi, A. 2013. What is inside the jar? Forensically Informative Nucleotide Sequencing (FINS) of a short mitochondrial COI gene fragment reveals a high percentage of mislabeling in jellyfish food products. Food Research International. 54(2): 1383-1393. https://doi.org/ 10.1016/j.foodres.2013.10.003
Armani, A., Tinacci, L., Xiong, X., Castigliego, L., Gianfaldoni, D., and Guidi, A. 2015. Fish species identification in canned pet food by BLAST and Forensically Informative Nucleotide Sequencing (FINS) analysis of short fragments of the mitochondrial 16S ribosomal RNA gene (16S rRNA). Food Control. 50: 821-830. https://doi.org/10.1016/j.food cont.2014.10.018
Clarke, S.C., Magnussen, J.E., Abercrombie, D.L., McAllister, M.K., and Shivji, M.S. 2006. Identification of shark species composition and proportion in the Hong Kong shark fin market based on molecular genetics and trade records. Conservation Biology. 20(1): 201-211.
Chen, C.H., Hsieh, C.H., and Hwang, D.F. 2012. Species identification of Cyprinidae fish in Taiwan by FINS and PCR-RFLP analysis. Food Control. 28(2): 240-245. https://doi.org/10.1016/j.foodcont.2012.05.012 Espineira, M., Vieites, J.M., and Santaclara, F. 2010. Species authentication of octopus, cuttlefish, bobtail and bottle squids (families Octopodidae, Sepiidae and Sepiolidae) by FINS methodology in seafoods. Food Chemistry. 121(2): 527-532. https://doi.org/10.1016/j.foodchem.2009.12.042
Fumagalli, L., Cabrita, C.J., and Castella, V. 2009. Simultaneous identification of multiple mammalian species from mixed forensic samples based on mtDNA control region length polymorphism. Forensic Science International: Genetics Supplement Series. 2(1): 302-303. https://doi.org/ 10.1016/j.fsigss.2009.08.009
Gupta, S.K., Thangaraj, K., and Singh, L. 2011. Identification of the source of ivory idol by DNA analysis. Journal of Forensic Sciences. 56(5): 1343-1345. https://doi.org/10.1111/j.1556-4029.2011.01750.x
Gupta, S.K., Kumar, A., Hussain, S.A., and Singh, L. 2013. Cytochrome b based genetic differentiation of Indian wild pig (Sus scrofa cristatus) and domestic pig (Sus scrofa domestica) and its use in wildlife forensics. Science and Justice. 53(2): 220-222. https://doi.org/10.1016/j.scijus.
2012.09.005
Hall, T.A. 1999. BiEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/95/NT. Nucleic Acids Symposium Series. 41: 95-98.
Hsieh, H.M., Chiang, H.I., Tsai, L.C., Lai, S.Y., Huang, N.E., Linacre, A., and Lee, J.C.I. 2001. Cytochrome b gene for species identification of the conservative animals. Forensic Science International. 122(1): 7-18. https://doi.org/10.1016/S0379-0738(01)00403-0
Hsieh, H.M., Huang, L.H., Tsai, L.C., Kuo, Y.C., Meng, H.H., Linacre, A., and Lee, J.C.I. 2003. Species identification of rhinoceros horns using cytochrome b gene. Forensic Science International. 136(1-3): 1-11. https://doi.org/10.1016/S0379-0738(03)00251-2
Imaizumi, K., Akutsu, T., Miyasaka, S., and Yoshino, M. 2007. Development of species identification tests targeting the 16S ribosomal RNA coding region in mitochondrial DNA. International Journal of Legal Medicine. 212: 184-191. https://doi.org/10.1007/s00414-006-0127-5
Iyengar, A. 2014. Forensic DNA analysis for animal protection and biodiversity conservation: A review. Journal of Nature Conservation. 22(3): 195-205. http://dx.doi.org/10.1016/j.jnc.2013.12.001
Johnson, R., Wilson-Wilde, L., and Linacre, A. 2014. Current and future directions of DNA in wildlife forensic science. Forensic Science International: Genetics. 10: 1-11. https://doi.org/10.1016/j.fsigen.2013.12.007
Jun, J., Han, S.H., Jeong, T.J., Park, H.C., Lee, B., and Kwak, M. 2011. Wildlife forensic using mitochondrial DNA sequences: Species identification based on hairs collected in the field and confiscated tanned Felidae leathers. Genes & Genomics. 33: 721-726. https://doi.org/ 10.1007/s13258-011-0080-7
Kitpipit, T., Tobe, S.S., Kitchener, A.C., Gill, P., and Linacre, A. 2012. The development and validation of a single SNaPshot multiplex for tiger species and subspecies identification-Implication for forensic purposes. Forensic Science International: Genetics. 6: 250-257. https://doi.org/ 10.1016/j.fsigen.2011.06.001
Kumar, U.S., Ratheesh, R.V., Thomas, G., and George, S. 2012. Use of DNA barcoding in wildlife forensics: a study of sambar deer (Rusa unicolor). Forest Science and Technology. 8: 224-226. https:// doi.org/10.1080/21580103.2012.750802
Lee, J.C., Tsai, L.C., Yang, C.Y., Liu, C.L., Huang, L.H., Linacre, A., and Hsieh, H.M. 2006. DNA profiling of Shahtoosh. Electrophoresis. 27: 3359-3362. https://doi.org/10.1002/elps.200600062
Li, M., Zhang, K.Y.B., But, P.P.H., and Shaw, P.C. 2011. Forensically informative nucleotide sequencing (FINS) for the authentication of Chinese medicinal materials. Chinese Medicine. 6: 42-47. https:// doi.org/10.1186/1749-8546-6-42
Librado, P., and Rozas, J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 25(11): 1451- 1452. https://doi.org/10.1093/bioinformatics/btp187
Lopez-Oceja, A., Gamarra, D., Borragan, S., Jimenez-Moreno, S., and de Pancorbo, M.M. 2016. New cyt b gene universal primer set for forensic analysis. Forensic Science International: Genetics. 23: 159-165. https://doi.org/10.1016/j.fsigen.2016.05.001
Parson, W., Pegoraro, K., Niederstatter, H., Foger, M., and Steinlechner, M. 2000. Species identification by means of the cytochrome b gene. International Journal of Legal Medicine.114(1-2): 23-28.
Rajapaksha, W., Thilakaratne, I., Chandrasiri, A., and Niroshan, T. 2003. Development of PCR assay for identification of buffalo meat. Asian-Australasian Journal of Animal Sciences. 16: 1046-1048. https://doi.org/10.5713/ajas.2003.1046
Sahajpal, V., and Goyal, S.P. 2010. Identification of a forensic case using microscopy and forensically informative nucleotide sequencing (FINS): a case study of small Indian civet (Viverricula indica). Science and Justice. 50: 94-97. https://doi.org/10.1016/j.scijus.2009.07.002
Santaclara, F.J., Perez-Martin, R.I., and Sotelo, C.G. 2014. Developed of a method for the genetic identification of ling species (Genypterus spp.) in seafood products by FINS methodology. Food Chemistry. 143: 22-26. https://doi.org/10.1016/j.foodchem.2013.06.004
Trivedi, S., Abdulhadi, A.A., Ansari, A.A., and Ghosh, S.K. 2016. Role of DNA barcoding in marine biodiversity assessment and conservation: an update. Saudi Journal of Biological Sciences. 23: 161-171. https:// doi.org/10.1016/j.sjbs.2015.01.001
Welton, L.J., Siler, C.D., Linkem, C.W., Diesmos, A.C., Diesmos, M.L., Sy, E., and Brown, R.M. 2013. Dragons in our midst: phyloforensics of illegally traded Southeast Asain monitor lizards. Biological Conservation. 159: 7-15. https://doi.org/10.1016/j.biocon.2012.10.013
Wen, J., Hu, C., Zhang, L., Luo, P., Zhao, Z., Fan, S., and Zu, T. 2010. The application of PCR-RFLP and FINS for species identification used in sea cucumbers (Aspidochirotida: Stichopodidae) products from the market. Food Control. 21: 403-407. https://doi.org/10.1016/j.foodcont.2009.06.014
Wilson-Wilde, L., Norman, J., Roberson, J., Sarre, S., and Georges, A. 2010. Current issue on species identification for forensic science and the validity of using cytochrome oxidase I (COI) gene. Forensic Science Medicine and Pathology. 6: 233-241. https://doi.org/10.1007/s12024- 010-9172-y
Thanat Vorajinda1, Chavin Chaisongkram2, Wibhu Kutanan3, and Khemika Lomthaisong1*
1Forensic Science Program, Faculty of Science, Khon Kaen University, Khon Kaen 40002, Thailand
2Zoological Organization Khon Kaen Zoo, 40002, Thailand
3Department of Biology, Faculty of Science, Khon Kaen University, Khon Kaen 40002, Thailand
*Corresponding author. E-mail: khemlo@kku.ac.th
Total Article Views
Article history:
Received: February 26, 2019;
Revised: March 27, 2019;
Accepted: April 5, 2019

