
Nutritional Requirements of Aeromonas sp. EBB-1 for Lipase Production
Pakamat Nualta and Jittima Charoenpanich*Published Date : 2019-08-25
DOI : 10.12982/CMUJNS.2014.0030
Journal Issues : Number 2,May-August-2014
ABSTRACT
The demand for the production of lipase, a widely used hydrolytic enzyme in biotechnological applications, has led to research on lipase-producing bacteria and culture strategies. In our previous study, we isolated Aeromonas sp. EBB-1 as a novel thermostable-lipase producer from marine sludge in Angsila, Thailand. Given this bacterium’s high production of lipase, we now further investigate the effects of different nutritional supplements on its lipase production. Maximum lipase activity (81-fold) was found after cultivating the strain in a medium containing 2.5% (w/v) yeast extract. Separately, adding 0.1% (w/v) glucose, 1.5% (w/v) proline and 0.5% (w/v) gum arabic enhanced lipase production activity nearly 9-, 10- and 6-fold, respectively. In contrast, adding a combination of nutrients in the same medium inhibited lipase production compared to the control. The result with yeast extract is especially promising, yielding high lipase concentrations from Aeromonas sp. EBB-1 in an inexpensive and simple medium.
Keywords: Nutritional sources, Aeromonas sp., Optimization, Lipase production, Thermostable lipase
INTRODUCTION
Lipases or triacylglycerol acylhydrolases (EC 3.1.1.3), one of the most versatile biocatalysts, are used in many biotechnological applications due to the different reactions they are able to catalyze and to their exquisite regio-specificity and chiral selectivity (Arbige and Pitcher, 1989; Jaeger et al., 1994 and Hasan et al., 2006). Most lipases used in industrial applications are generally distributed in plants, animals and microorganisms (Arbige and Pitcher, 1989; Jaeger et al., 1994; Fang et al., 2006). Among them, lipases of microbial origin are some of the most commonly used, since they can catalyze a variety of hydrolytic orsynthetic reactions (Jaeger and Reetz, 1998 and Schmid et al., 2001). The demand for the production of highly-active preparations of lipases has led to research on lipase-producing microorganisms and on culture strategies. Lipase production depends largely on culture practices and medium composition (Kim et al., 1996; Lotti et al., 1998; Dalmau et al., 2000). Finding a low-cost medium for the production of lipases would make even more industrial applications feasible.
In our laboratory, 22 strains of lipase-producing bacteria were previously isolated and screened for the production of thermostable lipase. Among them, Aeromonas sp. EBB-1 produced high lipase activity (6.50 ± 0.03 U/ml) and could be grown at high temperatures, of up to 55°C (Charoenpanich et al., 2011), which is attractive for industrial applications. This present study aims to improve lipase production of Aeromonas sp. EBB-1 by nutrient source optimization, in an effort to find the simplest and most economical medium.
MATERIALS AND METHODS
Bacterial strain and culture condition
The bacterial strain used in this study was isolated from marine sludge in Angsila, Thailand and identified as Aeromonas sp. EBB-1. This strain produces a thermostable lipase that is preferentially active towards long-carbon chain acylesters (Charoenpanich et al., 2011). Throughout the study, the general procedures for cultivation were as follows: 5.0 ml of 15 h bacterial inoculums was inoculated into 100 ml of 0.2 × Luria-Bertani (LB) medium (0.2% Bacto-tryptone, 0.2% NaCl and 0.1% Yeast extract, pH 7.2) containing analyzed supplement. Samples from the culture broths used in this study were taken from the late exponential phase of growth (OD600 ~ 0.8). Culture broth was centrifuged at 10,000 × g and 4°C for 20 min. The supernatant obtained was filtered through a 0.45 μm nylon membrane filter (Whatman, England) to collect the cell-free supernatant and use for enzymatic assay. Each experiment was done in triplicate.
Lipase activity assay and protein determination
Lipase activity was measured by a hydrolysis reaction using p-nitrophenyl palmitate as substrate (Sigma, Germany) according to the method of Pencreac’h and Baratii (1996). One unit (U) of enzyme was defined as the amount of enzyme releasing 1 μmol of p-nitrophenol per minute under the assay conditions. The amount of p-nitrophenol was calculated from the p-nitrophenol (Sigma, Germany) standard curve. Protein concentration was determined spectrophotometrically according to the method of Bradford (1976), using the Bio-Rad assay reagent (Hercules, USA) and bovine serum albumin as the standard. Statistical analysis was performed using a two-tailed t-test and P<0.05 was considered statistically significant.
Nutritional factors affecting lipase production by Aeromonas sp. EBB-1
To test the effect of different carbon sources on lipase production, various concentrations (0.05 - 0.2%) of carbon sources were added directly into the medium. All carbon sources were filter-sterilized by 0.2 μm nylon membrane filter (Whatman, England). The following carbon sources were studied: glucose, fructose, galactose, sucrose, glycerol and sugar.
The effect of nitrogen source (0.5-4.0% concentration) was investigated by adding directly into the medium. Organic nitrogen sources used in this study were tryptone, peptone, yeast extract and urea. Inorganic nitrogen sources were ammonium sulfate, potassium nitrate and ammonium nitrate.
For the amino acids experiment, amino acids (0.25-2.5% concentration) were added directly into the medium. The following amino acids were used: glycine, proline, arginine, aspartic acid, phenylalanine and cysteine.
The polysaccharides effect was determined by adding gum arabic, sodium alginate or agar, ranging in concentration from 0.5 to 1.0% (w/v). The hydrocarbon hexadecane was also used for investigation at the same concentrations.
RESULTS
Effect of carbon source on lipase production
In order to evaluate the effect of supplements on lipase production, various carbon sources at 0.1% (w/v) were tested preliminarily. The best carbon source for lipase production by Aeromonas sp. EBB-1 was glucose (66.24 ± 0.41 U/ml), with fructose (36.54 ± 1.61 U/ml) and galactose (24.55 ± 3.59 U/ml) yielding less lipase (Figure 1a). No significant difference in activity was found in the presence of sucrose, glycerol and sugar. The lipase yields obtained with either lower or higher concentrations of glucose were considerably less than those observed at 0.1% (w/v), as shown in Figure 1b.
Effect of nitrogen source on lipase production
The ability of Aeromonas sp. EBB-1 to produce lipase in medium was examined in different nitrogen sources (0.5% w/v). Maximum lipase production was noted in the yeast extract supplement (163.09 ± 1.94 U/ml). Others, in decreasing order of activity, were tryptone (74.94 ± 1.36 U/ml) and peptone (44.77 ± 1.86 U/ml) (Figure 2a). For inorganic nitrogen sources, urea and ammonium sulfate produced similarly low yields (20.85 ± 1.33 U/ml and 18.48 ± 1.40 U/ml, respectively). Interestingly, the addition of 2.5% (w/v) of yeast extract enhanced lipase production by 81-fold (529.69 ± 3.62 U/ml; Figure 2b).
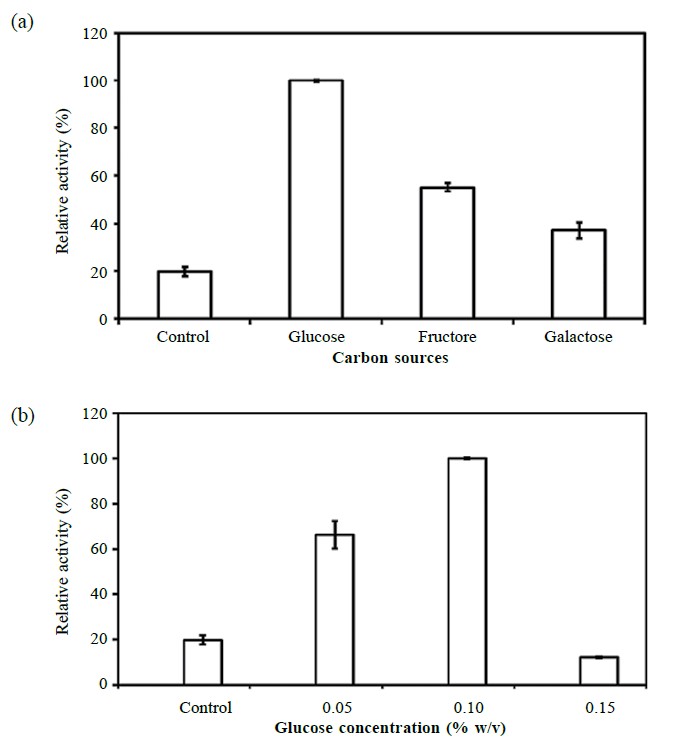
Figure 1. Effect of carbon sources (a) and concentration of glucose (b) on lipase production.
Note: Different carbon sources at 0.1% (w/v) were added to the 0.2 × LB medium and the strain cultivated at 25°C with continuous shaking at 250 rpm until the late exponential phase of growth. The relative activity was based on the highest lipase activity in the experiment and expressed as the mean of three determinations, with the standard deviations (mean ± SD) compared to 0.2 × LB medium (control).
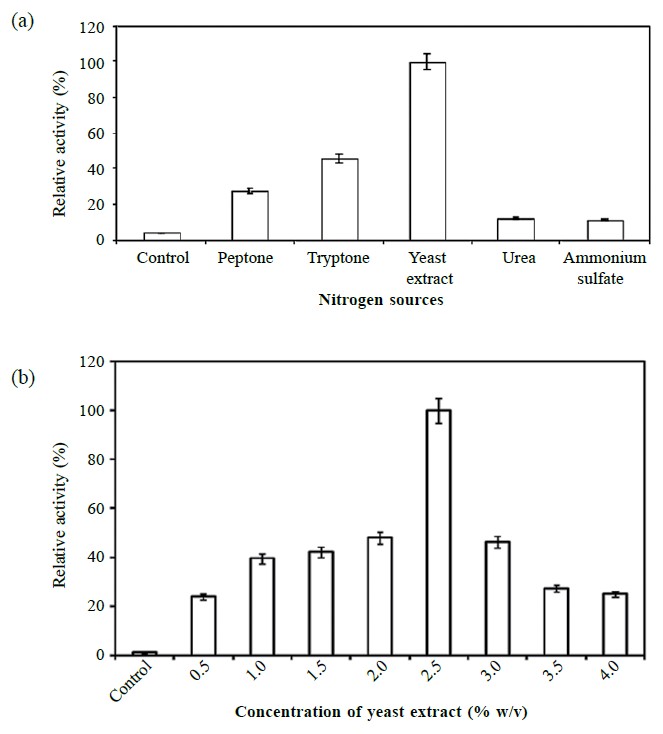
Figure 2. Effect of nitrogen sources (a) and concentration of yeast extract (b) on the production of lipase.
Note: Either organic or inorganic nitrogen source at 0.5% (w/v) was added to the 0.2 × LB medium and the strain cultivated at 25°C with continuous shaking at 250 rpm. The lipase production corresponds to those obtained at the late exponential phase of growth. These values are relative to the highest lipase activity in the experiment and expressed as the means ± SD (n = 3) compared to 0.2 × LB medium (control).
Effect of amino acid on lipase production
Experiments supplementing the medium with 0.5% (w/v) of amino acid were tried. Results indicated that the presence of arginine stimulated the highest lipase production (40.06 ± 2.74 U/ml), while proline (20.31 ± 2.08 U/ml), phenylalanine (17.62 ± 5.30 U/ml) and glycine (12.50 ± 1.11 U/ml) produced low yields, as shown in Figure 3a. Proline at a concentration of 1.5% (w/v, Figure 3b) enhanced lipase production nearly 10-fold, while 0.25% (w/v) arginine (Figure 3c) enhanced production ~6-fold.
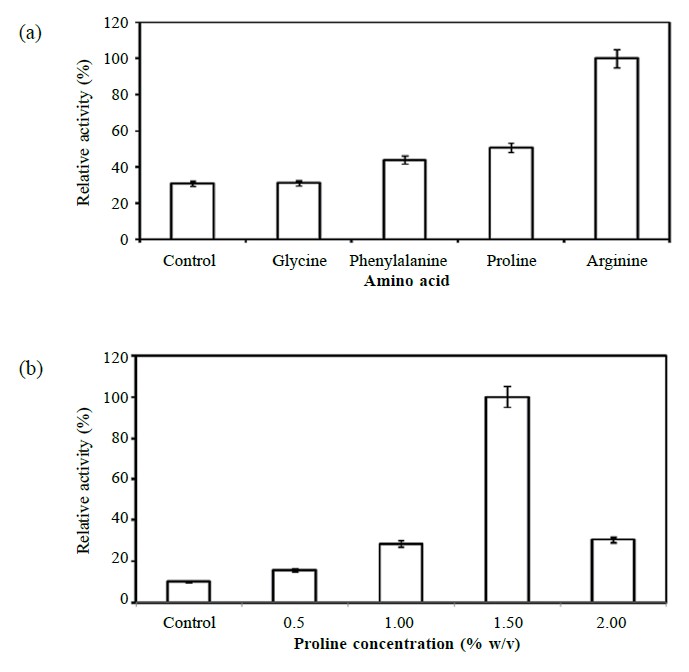
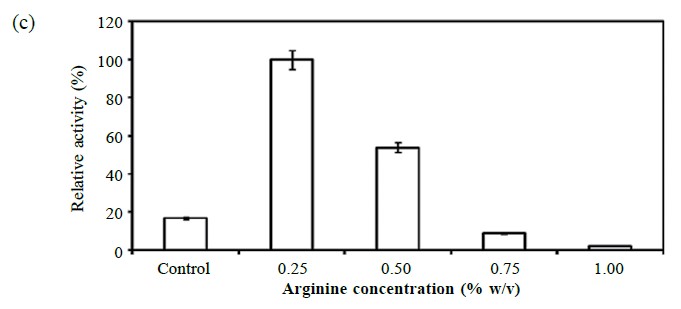
Figure 3. Effect of different amino acids (a) and concentrations of proline (b) and arginine (c) on lipase production.
Note: The samples were taken from the culture at the late exponential phase of growth (OD600 ~ 0.8). Experiments were done in triplicate and the activities of lipase were expressed as relative activity compared to the maximum (100% relative activity) observed value.
Effect of additives on lipase production
Three polysaccharides – gum arabic, agar and sodium alginate – were added separately to the medium at different concentrations. Among them, gum arabic produced the highest lipase activity (45.03 ± 5.07 U/ml), with the 0.5% (w/v) concentration promoting the maximum lipase production per unit of the growth of bacterium (Figure 4). On the other hand, sodium alginate and hexadecane showed inhibitory effects for lipase production. No significant production of lipase occurred when Aeromonas sp. EBB-1 was grown on the medium containing agar.
To complete these studies, the individual supplement giving the maximum lipase activity was chosen to evaluate its effect on lipase production (Figure 5). As expected, a mixture of yeast extract with other supplements produced higher lipase activity than the control. However, supplementing with yeast extract alone produced the highest activity.
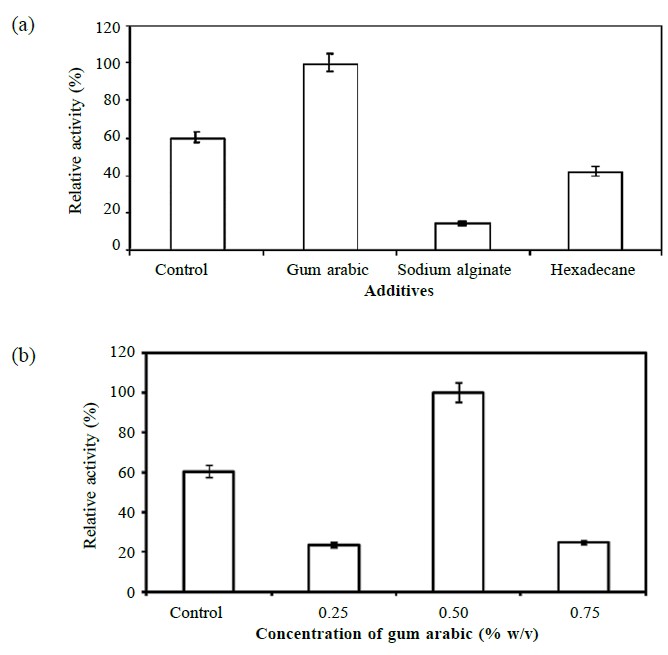
Figure 4. Effect of additives (a) and concentration of gum arabic (b) on lipase production.
Note: Different additives at 0.5% (w/v or v/v) were added to the 0.2 × LB medium and the strain cultivated until the late exponential phase of growth at 25°C with continuous shaking at 250 rpm. Media without the additives were used as control. Lipase activities were determined in triplicate and reported as averages ± standard deviation.
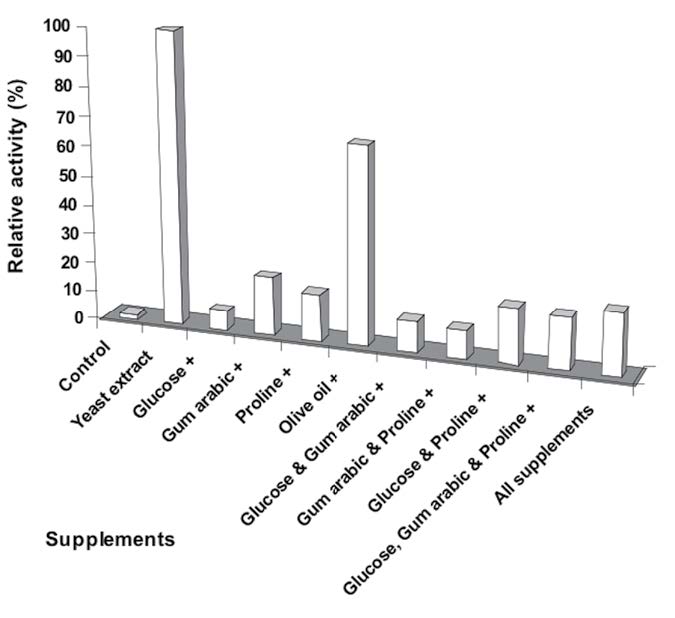
Figure 5. Relative activities among lipase produced by Aeromonas sp. EBB-1 using mixtures of nutritional supplements.
Note: Media without the additives were used as control. Lipase activities were determined in triplicates and averages ± SD reported.
DISCUSSION
Bacterial lipases are typically extracellular and influenced greatly by nutritional factors. In this study, we tried to find the optimum medium for lipase production of Aeromonas sp. EBB-1, a novel thermostable lipase isolated in a previous study (Charoenpanich et al., 2011). It has been reported that monosaccharides, especially glucose, stimulated both enzyme production in different microorganisms and secretion of lipase accumulated inside the cells (Macfarlane and Macfarlane, 1992; Mehrotra et al., 1999; Dalmau et al., 2000; Boekema et al., 2007 and Uttatree and Charoenpanich, 2011). On the other hand, the repression of enzyme synthesis in the liquid medium by sucrose and other readily-metabolized carbon sources was referred to as catabolite repression, the paradigm of cellular regulation for the low preferential carbon source (Stülke and Hillen, 1999; Brückner and Titgemeyer, 2002; Deutscher, 2008 and Uttatree and Charoenpanich, 2011). In the presence of disaccharides, for example sucrose, catabolite repression in Aeromonas sp. EBB-1 might serve as an autoregulatory device to keep sucrose utilization at a certain level, leading to the lower production of lipase rather than to establish preferential utilization of glucose.
In most microorganisms, either inorganic or organic nitrogen sources are metabolized to produce amino acids, nucleic acids, proteins and cell wall components. The results suggest that inorganic nitrogen sources are not essential for growth and lipase production by this strain, although the growth and enzyme productivity were low. The lipase yields obtained with potassium nitrate and ammonium nitrate were considerably lower than the control. This phenomenon might be due to the repression of enzyme synthesis by the rapidly metabolizable ammonium ion concentration in the medium, which interfered with the utilization and metabolism of peptides through catabolite repression (Giesecke et al., 1991 and Snowden et al., 1992). Moreover, both sulfate and nitrate affects calcium availability by precipitation or chelation and, therefore, reduces lipase production (Freire et al., 1997.). Interestingly, the addition of 2.5% (w/v) of yeast extract enhanced lipase production by 81-fold (529.69 ± 3.62 U/ml), even higher than with an olive oil supplement (440 ± 13 U/ml) described previously (Charoenpanich et al., 2011). The simple addition of yeast extract into the medium resulted in higher lipase production of Aeromonas sp. EBB-1 than with the oil supplement.
Incorporation of amino acids in the production medium might enhance lipase production (Arbige and Pitcher, 1989; Bornscheuer et al., 2002). At a concentration
of 0.5% (w/v), arginine stimulated the highest lipase production, but others produced low yields. Proline at a concentration of 1.5% (w/v) enhanced lipase production nearly 10-fold, while 0.25% (w/v) arginine enhanced production approximately 6-fold. The inhibitory effect of glycine has been reported previously (Ikura and Horikoshi, 1987; Uttatree and Charoenpanich, 2011).
The increase in lipase production by adding gum arabic might be due to this compound enhancing the mechanical liberation of the enzyme at the cell surface (Winkler and Stuckmann, 1979; Mahler et al., 2000; Martinez and Nudel, 2002). A combination of yeast extract with other supplements produced higher lipase activity than the control, but still lower than supplementing with yeast extract alone. This is important; proving that yeast extract alone provided a simple and convenient nitrogen source for industrial-scale lipase production.
By optimization of medium components, the production of an extracellular lipase from Aeromonas sp. EBB-1 was improved. After addition of yeast extract (2.5% w/v), an 81-fold increase of lipolytic activity was found compared to that obtained using 0.2 × LB medium. Adding 0.1% (w/v) glucose, 1.5% (w/v) proline and 0.5% (w/v) gum arabic also improved lipase production, although to a lesser extent (6-10 fold). However, adding a combination of nutrients in the same medium inhibited lipase production compared to the control. These results are promising because this strain produces lipases in a simple medium (0.2% Bacto-tryptone, 0.2% NaCl and 2.6% Yeast extract, pH 7.2).
ACKNOWLEDGEMENT
This work was partially supported by Senior Project Funding, Faculty of Science, Burapha University.
REFERENCES
Arbige, M.V., and W.H. Pitcher. 1989. Industrial enzymology: a look towards the future. Trends in Biotechnology 7: 330-335.
Boekema, B.K.H.L., A. Beselin, M. Breuer, B. Hauer, M. Koster, F. Rosenau, K.E. Jaeger, and J. Tommassen. 2007. Hexadecane and Tween 80 stimulate lipase production in Burkholderia glumae by different mechanisms. Applied and Environmental Microbiology 73: 3838-3844. 10.1128/AEM.00097-07
Bornscheuer, U.T., C. Bessler, R. Srinivas, and S.H. Krishna. 2002. Optimizing lipase and related enzymes for efficient application. Trends in Biotechnology 20: 433-437. 10.1016/S0167-7799(02)02046-2
Bradford, M.M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing, the principle of protein-dye binding. Analytical Biochemistry 72: 248-254.
Brückner, R., and F. Titgemeyer. 2002. Carbon catabolite repression in bacteria: choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiology Letters 209: 141-148. 10.1016/S0378-1097(02)00559-1
Charoenpanich, J., S. Suktanarag, and N. Toobbucha. 2011. Production of a thermostable lipase by Aeromonas sp. EBB-1 isolated from marine sludge in Angsila, Thailand. ScienceAsia 37: 105-114. 10.2306/scienceasia1513-1874.2011.37.105
Dalmau, E., J.L. Montesinos, M. Lotti, and C. Casas. 2000. Effect of different carbon sources on lipase production by Candida rugosa. Enzyme and Microbial Technology 26: 657-663. 10.1016/S0141-0229(00)000156-3
Deutscher, J. 2008. The mechanisms of carbon catabolite repression in bacteria. Current Opinion in Microbiology 11: 87-93. 10.1016/j.mib.2008.02.007
Fang, Y., Z. Lu, F. Lv, X. Bie, S. Liu, Z. Ding, and W. Xu. 2006. A newly isolated organic solvent tolerant Staphylococcus saprophyticus M36 produced organic solvent-stable lipase. Current Microbiology 53: 510-515. 10.1007/S00284-006-0260-X
Freire, D.M.G., E.M.F. Teles, E.P.S. Bon, and G.L. San’t Anna Jr. 1997. Lipase production by Penicillium restrictum in a bench-scale fermenter: Effect of carbon and nitrogen nutrition, agitation and aeration. Applied Biochemistry and Biotechnology 63: 409-421.
Giesecke, U.E., G. Bierbaum, H. Rudde, U. Spohn, and C. Wandrey. 1991. Production of alkaline protease with Bacillus licheniformis in controlled fed-batch process. Applied Microbiology and Biotechnology 35: 720-724.
Hasan, F., A.A. Shah, and A. Hameed. 2006. Industrial applications of microbial lipases. Enzyme and Microbial Technology 39: 235-251. 10.1016/j.enzmicte.2005.10.016
Ikura, Y., and K. Horikoshi. 1987. Effect of amino compounds on alkaline amylase production by alkaliphilic Bacillus sp. Journal of Fermentation Technology 65: 707-709.
Jaeger, K.E., S. Ransac, B.W. Dijkstra, C.C. van Henrel, and O. Misset. 1994. Bacterial lipases. FEMS Microbiology Reviews 15: 29-63. 10.1016/0168-6445(94)90025-6
Jaeger, K.E., and M.T. Reetz. 1998. Microbial lipases from versatile tools for biotechnology. Trends in Biotechnology 16: 396-403. 10.1016/S0167-7799(98)01195-0
Kim, S.S., E.K. Kim, and J.S. Rhee. 1996. Effects of growth rate on the production of Pseudomonas fluorescens lipase during the fed-batch cultivation of Escherichia coli. Biotechnology Progress 12(5): 718-722.
Lotti, M., S. Monticelli, J.L. Montesinos, S. Brocca, F. Valero, and J. Lafuente. 1998. Physiological control on the expression and secretion of Candida rugosa lipase. Chemical and Physical Lipids 93: 143-148. 10.1016/S0009-3084(98)00038-3
Macfarlane, G.T., and S. Macfarlane. 1992. Physiological and nutritional factors affecting synthesis of extracellular metalloproteinase by Clostridium bifermentans.
Applied and Environmental Microbiology 58: 1973-1976.
Mahler, G.F., R.G. Kok, A. Cordenons, K.J. Hellingwerf, and B.C. Nudel. 2000. Effects of carbon sources on extracellular lipase production and lipA transcription in Acinetobacter calcoaceticus. Journal of Industrial Microbiology and Biotechnology 24: 25-30.
Martinez, D.A., and B.C. Nudel. 2002. The improvement of lipase secretion and stability by addition of inert compounds into Acinetobacter calcoaceticus cultures. Canadian Journal of Microbiology 48: 1056-1061. 10.1139/W02-108
Mehrotra, S., P.K. Pandey, R. Gaur, and N.S. Darmwal. 1999. The production of alkaline protease by a Bacillus species isolate. Bioresource Technology 67: 201-203. 10.1016/S0960-8524(98)00107-2
Pencreac’h, G., and J.C. Baratti. 1996. Hydrolysis of p-nitrophenyl palmitate in n-heptane by the Pseudomonas cepacia lipase: A simple test for the determination of lipase activity in organic media. Enzyme and Microbial Technology 18: 417-422. 10.1016/0141-0229(95)00120-4
Schmid, A., J.S. Dordick, B. Hauer, A. Kiener, M. Wubbolts, and B. Witholt. 2001. Industrial biocatalysis today and tomorrow. Nature 409: 258-268. 10.1038/35051736
Snowden, L.Y., I.I. Blumentals, and R.M. Kelly. 1992. Regulation of proteolytic activity in the hyperthermophiles Pyrococcus furisosus. Applied and Environmental Microbiology 58: 1134-1141.
Stülke, J., and W. Hillen. 1999. Carbon catabolite repression in bacteria. Current Opinion in Microbiology 2: 195-201. 10.1016/S1369-5274(99)80034-4
Uttatree, S., and J. Charoenpanich. 2011. Nutritional requirements and physical factors affecting the production of organic solvent-stable lipase by Acinetobacter baylyi. Chiang Mai University Journal of Natural Sciences 10(1): 115-132.
Winkler, U.K., and M. Stuckmann. 1979. Glycogen, hyaluronidate and some other polysaccharides greatly enhance the formation of exocellular lipase by Serratia marcescens. Journal of Bacteriology 138: 663-670.
Pakamat Nualta1 and Jittima Charoenpanich1,2,3*
1 Department of Biochemistry and Centre of Excellence for Innovation in Chemistry (PERCH-CIC), Faculty of Science, Burapha University, Bangsaen, Chonburi 20131, Thailand
2 Environmental Science Program, Faculty of Science, Burapha University, Bangsaen, Chonburi 20131, Thailand
3 Centre of Excellence on Environmental Health and Toxicology (CHE), Ministry of Education, Bangkok 10400, Thailand
*Corresponding author. E-mail: jittima@buu.ac.th
Total Article Views

