
Proteome Analysis of Acrylamide-induced Proteins in a Novel Acrylamide-degrader Enterobacter aerogenes by 2D Electrophoresis and MALDI-TOF-MS
Jittima Charoenpanich* and Akio TaniPublished Date : 2019-08-24
DOI : 10.12982/cmujns.2014.0016
Journal Issues : Number 1, January-april 2014
ABSTRACT
Despite tremendous advances in understanding the microbial degradation of acrylamide, reports on the nature of the two-dimensional protein patterns for acrylamide-degrading bacteria are not yet available. This work, focusing on the acrylamide-inducible proteins, studied the response of Enterobacter aerogenes, a novel acrylamide-degrading bacterium, to acrylamide. Proteome analysis was applied using 2D-polyacrylamide gel electrophoresis and matrixassisted laser desorption/ionisation-time of flight mass spectrometry to identify proteins differentially expressed from E. aerogenes grown on acrylamide. Six protein homologues with amidohydrolase, urease accessory protein, quaternary ammonium compound resistance proteins, dipeptide transport protein, Omp36 osmoporin and large conductance mechanosensitive channel proteins (MscL) are seemingly involved in acrylamide stress response and its degradation. Five proteins identified as GroEL-like chaperonin, ArsR-transcriptional regulator, Ts- and Tu-elongation factor and trigger factor and four proteins (phosphoglycerate kinase, ATP synthase β-subunit, malate dehydrogenase and succinyl-CoA synthetase α-subunit) responsive for the adaption of E. aerogenes in the presence of acrylamide
Keywords: Acrylamide, Biodegradation, Enterobacter aerogenes, 2D-PAGE, MALDI-TOF MS
INTRODUCTION
Acrylamide (CH2=CHCONH2) is used as a conjugated reactive molecule in polyacrylamide production as well as a binding, thickening or flocculating agent in industrial applications (Prasad, 1982; Wampler and Ensign, 2005; Prabu and Thatheyus, 2007). Demand for acrylamide increases with industrial and domestic applications for polyacrylamides and other polymers (Prasad, 1982; Nagasawa and Yamada, 1989; Wang et al., 2009). However, acrylamide is highly neurotoxic, carcinogenic and teratogenic in animals and considered an environmental contaminant (Cherry et al., 1956; Croll et al., 1974; Tilson and Cabe, 1979; IARC, 1994; Segerbäck et al., 1995; Prabu and Thatheyus, 2007). Bioremediation of acrylamide to non-harmful substances would alleviate environmental concerns.
Microbial degradation of acrylamide has been explored extensively with a diversity of isolates, such as Bacillus, Pseudomonas and Rhodococcus (Yamada et al., 1979; Thiery et al., 1986; Nawaz et al., 1993; Hirrlinger et al., 1996; Wang and Lee, 2001; Shukor et al., 2009a, b). Acrylamide is initially deamidated to ammonia and acrylic acid, a process catalyzed by amidase or amidohydrolase (EC 3.5.1.4) (Shanker et al., 1990; Nawaz et al., 1994; Nawaz et al., 1998; Zabaznaya et al., 1998). Wampler and Ensign (2005) proposed acrylic acid is degraded initially to β-hydroxypropionate, then oxidized to CO2 or reduced to propionate.
In a previous study, a novel acrylamide-degrading bacterium, Enterobacter aerogenes was isolated and characterized from domestic wastewater in Chonburi, Thailand. The strain grew well in acrylamide as 0.5% (w/v), at pH 6.0 to 9.0 and 25°C, that was degraded to acrylic acid in the late logarithmic growth phase in a biomass-dependent pattern. Activity in cell-free supernatant was sufficient to completely degrade butyramide and urea, and other aliphatic amides to acrylic acid (Buranasilp and Charoenpanich, 2011). However, it remains unknown how acrylamide enter bacteria and the degradation process after its early conversion to acrylic acid. Other proteins may be involved in acrylamide and acrylic acid degradation. Cellular proteins can be separated by two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) (Klose, 1975; O’Farrell, 1975) and their apparent molecular weight and quantity measured allowing identification and is a powerful tool to study changes in cellular protein expression (Choe and Lee, 2000). The objective of the present study was to document the entire protein complement induced by acrylamide, using a combination of 2D-PAGE and MALDI-TOF mass spectrometry.
MATERIALS AND METHODS
Cell cultivation and harvesting
E. aerogenes (Buranasilp and Charoenpanich, 2011) was cultivated at 25°C in W-minimum medium (Kimbara et al., 1989) that contained 0.5% (w/v) acrylamide or glucose until the late exponential phase (OD600 ~ 1.0). Cells were harvested by centrifugation at 5,000 ×g for 10 min at 4°C, washed with 0.85% (w/v) NaCl, and stored at -80°C until use.
Preparation of protein fraction
Cell pellets were resuspended in 5-ml of lysis buffer I (10 mM Tris-HCl, 1 mM EDTA at pH 8.0, 1 mM PMSF and 1 mM DTT) and disrupted by ultrasonica tion (Buranasilp and Charoenpanich, 2011). Nuclease solution (1 mg/ml DNaseI, 0.25 mg/ml RNaseA, 50 mmol/l MgCl2, 24 mM Tris (base) and 476 mM Tris-HCl) at 1/10 volume was added and the mixture incubated on ice (<4°C) for 20 min to remove nucleic acid contaminants. Intact cells were removed by centrifugation at 10,000 ×g for 30 min, <4°C. The supernatant was collected by centrifugation at 40,000 ×g for 45 min (4°C), precipitated in 9 volumes of acetone solution I (10% (w/v) TCA and 0.3% (w/v) DTT in acetone) and kept overnight at -20°C. Proteins in the precipitate were collected by centrifugation at 5000 ×g for 5 min and washed with an appropriate volume of acetone solution II (0.3% (w/v) DTT in acetone). Washed proteins were then dissolved for 30 min in solubilization buffer (8 mM urea, 4% (w/v) CHAPS, 60 mM DTT, 2% (v/v) Bio-Lyte pH 3-10 (Bio-Rad, USA), and 0.002% (w/v) bromophenol blue). Insoluble components and reagent contaminants were removed by centrifugation at 15,000 ×g for 15 min. Dissolved proteins were determined spectrophotometrically (Bradford, 1976) using Bio-Rad assay reagent (Hercules, USA) and bovine serum albumin as the standard.
Two-dimensional electrophoresis
An immobilized gel strip (18 cm) containing a pH gradient of 5-8 (IPG) (Bio-Rad, USA) was rehydrated at 50 V, 20°C for 12 h with 350 μl of rehydration solution (8M urea, 0.5% (w/v) CHAPS, 0.2% (w/v) DTT, 0.5% (v/v) Bio-Lytes pH 3-10, and 0.002% (w/v) bromophenol blue) that contained 1 μg of protein sample. IEF was conducted at 20°C as follows: S1 – rapid voltage at 100 V for 1 h; S2 – rapid voltage at 500 V for 1 h; S3 – rapid voltage at 10,000 V for 10 h; S4 – rapid voltage from 10,000 to 60,000 V for 16 h; and S5 – rapid voltage at 500 V for 20 min. The limit voltage was 50 μA/gel strip. After focusing, gel strips were equilibrated first in solution I (50 mM Tris-HCl at pH 8.8, 6M urea, 30% (v/v) glycerol, 2% (w/v) SDS, 2% (w/v) DTT, and 0.002% (w/v) bromophenol blue), followed by solution II (50 mM Tris-HCl at pH 8.8, 6M urea, 30% (v/v) glycerol, 2% (w/v) SDS, 2.5% (w/v) iodoacetamide, and 0.002% (w/v) bromophenol blue) with gentle shaking for 10 min. Then, the IPG was embedded in a discontinuous SDS-PAGE gel (Laemmli, 1970) (15% separating gel, 5% stacking gel). Electrophoresis buffer consisted of 25 mM Tris-HCl at pH 8.3, 192 mM glycine, and 0.1% (w/v) SDS. Gels were run at constant current as 20 mA per gel until bromophenol blue moved into the separating gel, and then the current was raised to 50 mA per gel until complete. The gels were fixed in prefixative solution (20% (v/v) methanol and 7.5% (v/v) acetic acid) for 30 min, stained (50% (v/v) methanol, 10% (v/v) acetic acid, and 0.05% Coomassie brilliant blue R-250) for 45 min, and destained (5% (v/v) methanol and 7% (v/v) acetic acid) until the gel background was clear.
In-gel digestion and MALDI-TOF MS analysis
Protein spots from cells grown in acrylamide and glucose were compared, excised from 2-D gels and digested in trypsin (MS-grade, Sigma, USA) according to the proteomic protocols for mass spectrometry (Bruker Daltonics). Samples were concentrated and purified by ZipTipC18 resin (Millipore, England). Samples (0.8 μl) were then spotted on an AnchorChip target (Bruker, Germany) and analyzed with an UltraFLEX MALDI-TOF mass spectrometer (Bruker, Germany). Calibration was done with a peptide calibration standard, molecular mass 1 to 4 kDa (Bruker, Germany). The matrix solution was 0.3 mg/ml α-cyano-4-hydroxycinnamic acid (Bruker, Germany) in 50% acetone in ethanol. Trypsin contamination peak was excluded from the database peak list. Each spectrum was produced by accumulating data from 200 consecutive laser shots. Parent masses were analyzed with Biotools version 2.2 (Bruker, Germany) and amino acid sequences were identified using homology searches in the MS database (http://dove.embl-heidelberg.de/Blast2/msblast.html). Search parameters were as follows: type of search – peptide mass fingerprint; enzyme – trypsin; fixed modification – carbamido methylation (Cys); variable modifications – oxidation (Met); mass values – monoisotopic; peptide charge state – 1+; maximum missed cleavages – 1; and peptide mass tolerance – 0.05% Da (50 ppm). Statistical analyses of the sequences were determined by the probability-based Mowse score offered by the software. A p-value of < 0.05 was considered significant and used to generate the results.
RESULTS
In media from acrylamide-grown bacteria, 15 proteins were separated on 2D-gel run with a pH gradient of 5 to 8 with molecular sizes of approximately 56 (pI 5.7), 59 (pI 5.9), 41 (pI 5.3), 42 (pI 5.5), 60 (pI 5.4), 46 (pI 5.4), 15 (pI 5.5), 25 (pI 5.8), 11 (pI 6.2), 30 (pI 6.3), 33 (pI 6.3), 12 (pI 5.6), 48 (pI 7.1), 43 (pI 7.2), and 30 (pI 7.1) respectively (Fig. 1). Spots were cut from gels and subsequently digested in trypsin. Consequently, the digestate was analyzed by MALDI-TOF MS to determine molecular masses of the tryptic peptides. Sequences were compared with those in databases and potential identities are summarized in Table 1.
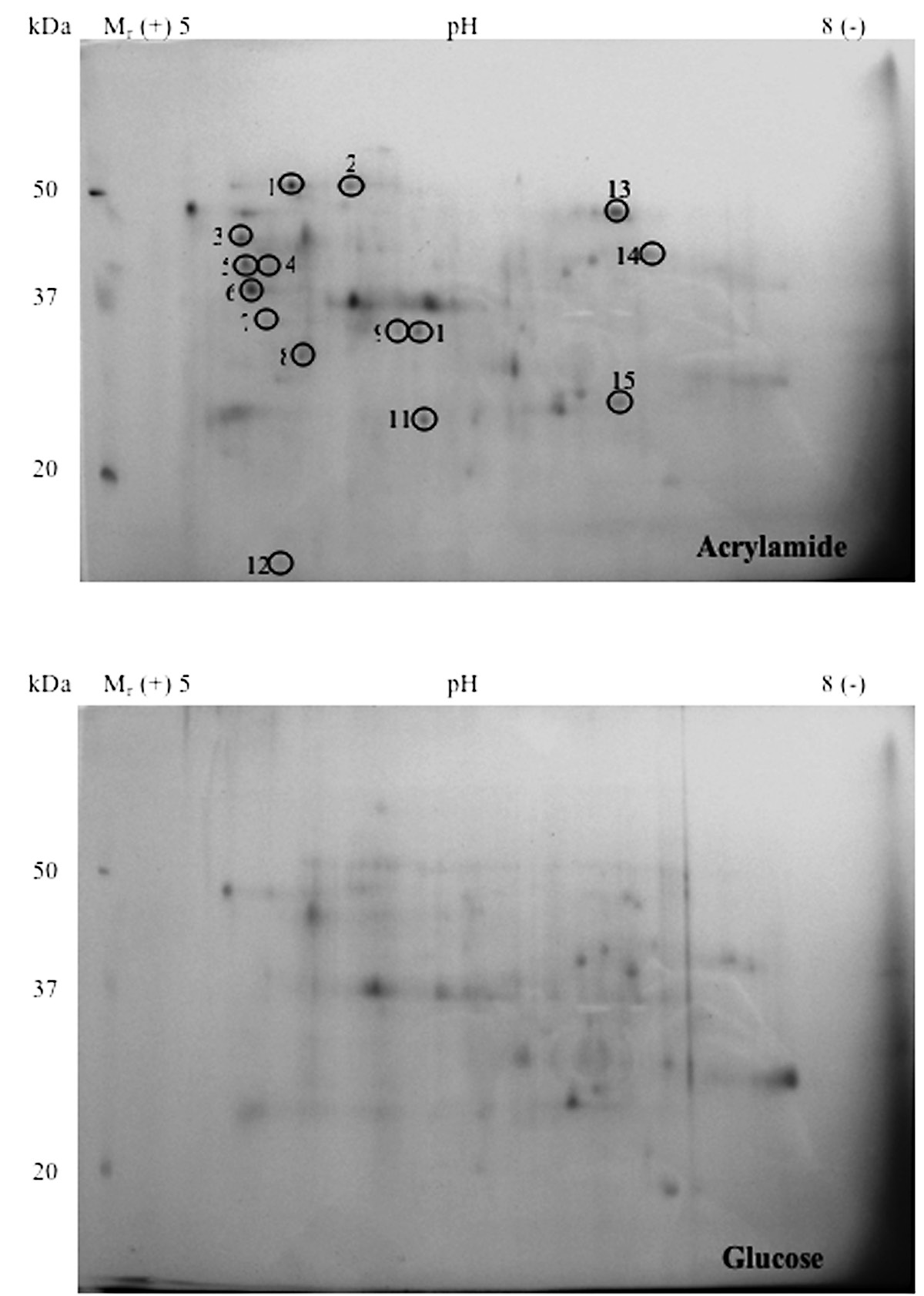
Figure 1. Proteins in (a) acrylamide- (b) glucose-grown E. aerogenes separated by 2D-gel electrophoresis using pH 5-8 IPG and 15% SDS-PAGE. Proteins were stained with Coomassie brilliant blue R-250. Different protein spots are circled and the identified proteins are labeled in Table 1.
Table 1. Summary of proteins identified from the 2D-gel of acrylamide-induced protein in E. aerogenes, compared with the cells grown on glucose and acrylamide.
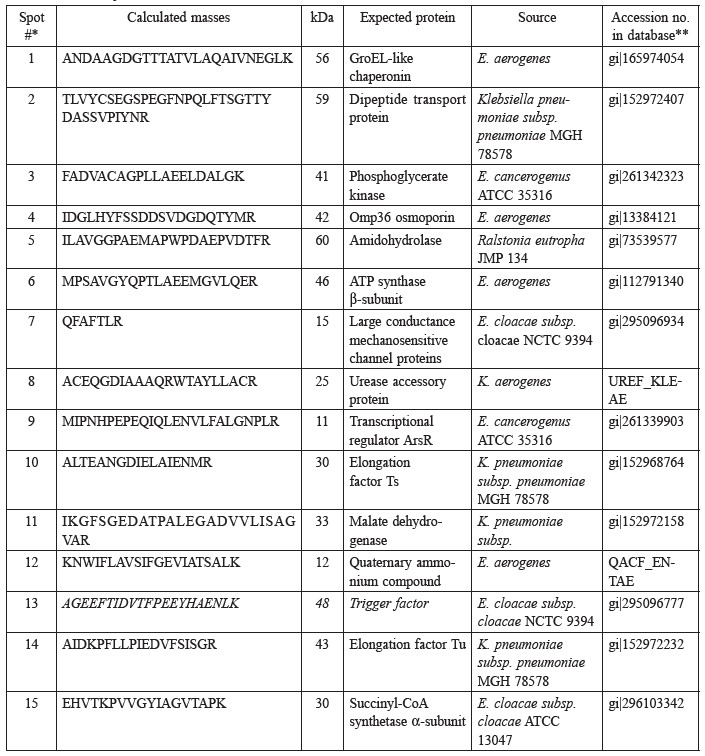
Note: *The spot numbers listed in the first column correspond to the spot numbers in the 2D-gel in Fig. 1. **MS database (http://dove/embl-heidelberg.de/Blast2/msblast.html).
Enzymes involved in energy metabolism are represented by four protein spots (phosphoglycerate kinase [Spot #3], ATP synthase β-subunit [Spot #6], malate dehydrogenase [Spot #11], succinyl CoA synthase α-subunit [Spot #15]) and five, in the synthesis and transport of proteins (GroEL-like chaperonin [Spot #1], transcription regulator ArsR [Spot #9], elongation factor Ts [Spot #10] and Tu [Spot #14], trigger factor [Spot #13]). The acrylamide stress response may also involve the gel-abundant dipeptide transport proteins [Spot #2], Omp 36 osmoporin [Spot #4], amidohydrolase [Spot #5], large conductance mechanosensitive channel protein [MscL, Spot #7], urease accessory protein [Spot #8] and quaternary ammonium compound resistance protein [Spot #12].
DISCUSSION
Until now, we cannot deny possible routes for acrylamide other than deamination via amidase (Nawaz et al., 1994; Hirrlinger et al., 1996; Cha & Chambliss, 2011). The subsequent fate of acrylate is not well understood, but probably involves pathways and enzymes that have been characterized to various degrees for other acrylate-utilizing bacteria. Acrylate metabolism is believed to proceed via hydroxylation to β-hydroxypropionate, then oxidized to CO2 (Shanker et al., 1990) or reduced to propionate (Wampler and Ensign, 2005). Another plausible pathway for mineralization of acrylamide is via formation of acrylyl CoA, which eliminates lactate as a final product (Shanker et al., 1990).
A powerful tool that also enables unraveling acrylamide metabolic pathways is the sequential induction of catabolic enzymes and intermediatary metabolites. Further, insight into degradative pathways is also provided from assaying the probable key proteins that are synthesized at sufficient levels when acrylamide is present. Using proteome analysis, fifteen proteins differentially expressed from Enterobacter aerogenes grown on acrylamide were identified. Five proteins identified as GroEL-like chaperonin, ArsR-transcriptional regulator, Ts- and Tuelongation factor and trigger factor and four proteins (phosphoglycerate kinase, ATP synthase β-subunit, malate dehydrogenase and succinyl-CoA synthetase α-subunit) are expected to be relevant to adaption of E. aerogenes in the presence of acrylamide. The metabolic changes may be a consequence of the biological cost of acrylamide resistance, although other forms of these proteins may exist and are not resolved on these 2D-gels. Although chaperonin provides essential kinetic assistance to protein folding, expression of this protein for cell survival in nutrient limitations have also been documented (Fayet et al., 1989; Fischer et al., 1993). Chaperonin may be required for stress-induced survival in acrylamide. ArsR transcription regulator implicitly represents a detoxification mechanism that has endowed prokaryotes to respond to externally induced stress. Most appear to be negative regulators, and many derepress by direct binding of metal ions (Busenlehner et al., 2003). Thus, and perhaps most interesting, this regulator might be necessary for expression of genes relevant in acrylamide metabolism. However, the functions of this regulator in acrylamide degradation are to be elucidated. Elongation factor Ts and Tu are expected to be the essential proteins for transcription and translation of proteins involved in acrylamide degradation.
Six protein homologues with amidohydrolase, urease accessory protein, quaternary ammonium compound resistance proteins, dipeptide transport protein, Omp36 osmoporin and large conductance mechanosensitive channel proteins (MscL) are seemingly involved in acrylamide stress response and its degradation. The outer membrane of gram-negative bacterial cells forms a protective barrier against damaging external agents. Porins in the membrane form hydrophilic channels, allowing selective uptake of essential nutrients (Koebnik et al., 2000; Nikaido, 2003). Synthesis of porins may be up- or down-regulated by the presence or absence of molecules in the medium or inducer threshold concentrations (Barbosa and Levy, 2000; Pomposiello et al., 2001; Nikaido, 2003). Perhaps E. aerogenes uses Omp36 osmoporin to assimilate acrylamide. Systems to transport peptides occur in many species and generally function to accumulate intact peptides intracellularly, where they are hydrolyzed (Payne, 1980). The dipeptide transport protein, one of the osmotic-shockable transport systems of gram-negative bacteria is responsible for the uptake of many types of small molecules (Payne, 1980; Payne and Smith, 1994). As an amide, the movement of acrylamide from periplasm into the cell of E. aerogenes might be facilitated by dipeptide transport protein
The present study confirms the production of amidohydrolase (amidase) by E. aerogenes during acrylamide degradation and is in accord with earlier reports (Shanker et al., 1990; Nawaz et al., 1994; Nawaz et al., 1998; Buranasilp and Charoenpanich, 2011; Thanyacharoen et al., 2012). MscL protein of the mechanosensitive channel with large conductance appears to play an important role in the integrity of cell membranes by regulating the flow of ions and other small solutes (Ajouz et al., 1998; Berrier et al., 2000). Thus, MscL might release ammonium or other ions from E. aerogenes for cell stabilization. This hypothesis is confirmed by the release of ammonium ion into the media of E. aerogenes (Buranasilp and Charoenpanich, 2011). The functions of quaternary ammonium compound resistance protein as well as urease are still ambiguous but may be involved in ammonium detoxification.
Although enzyme assays and other physiological data on E. aerogenes are still needed, the present study suggests acrylamide may be assimilated using Omp36 osmoporin and dipeptide transport proteins (Figure 2).
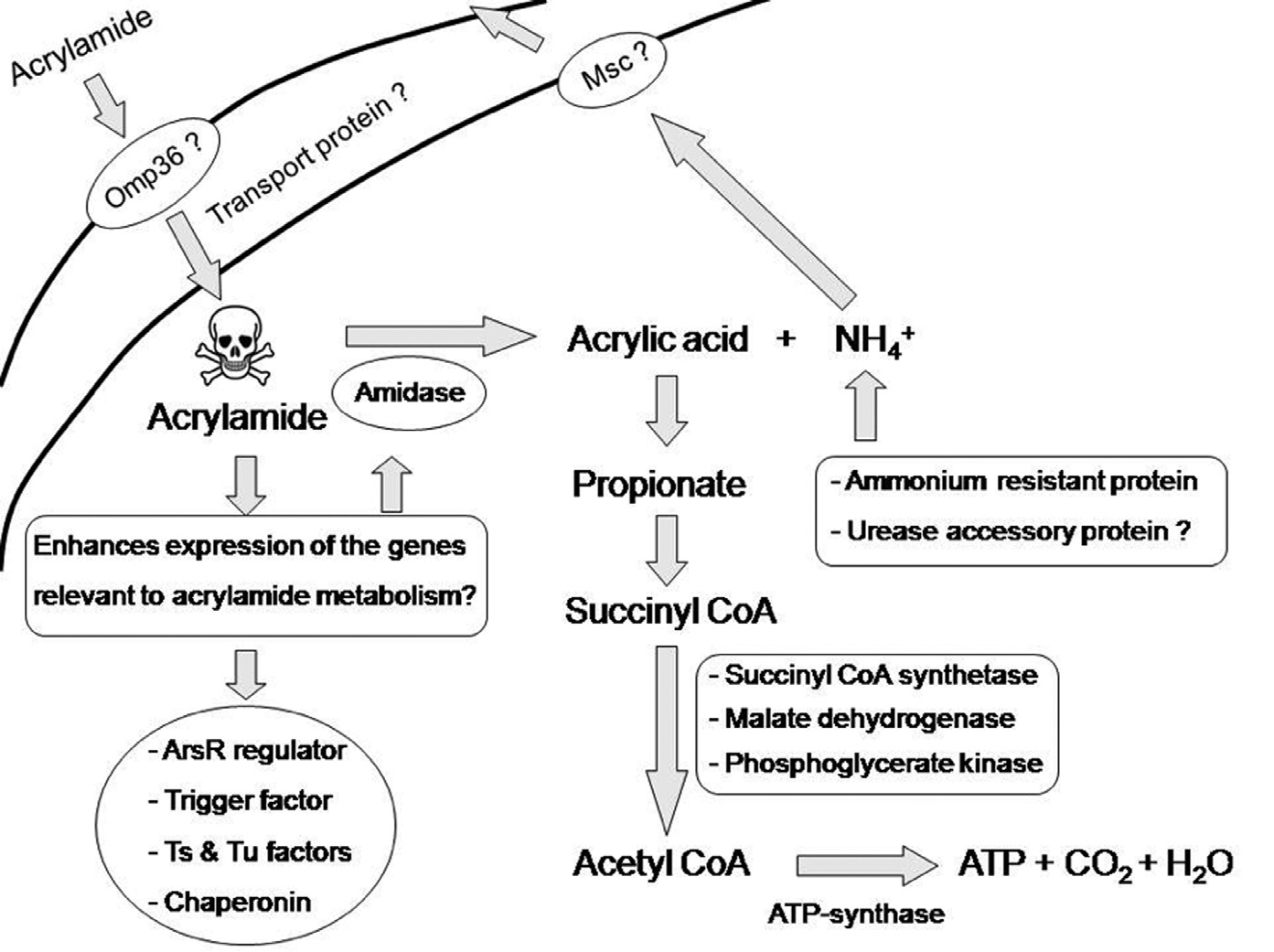
Figure 2. Model of acrylamide uptaking and the subsequent fate of acrylamide possible occurred in E. aerogenes. Details are given in the text.
Acrylamide is toxic, indeed lethal, to most microorganisms. However some bacteria, including E. aerogenes, have adapted their metabolism to use this substance as an energy source. Important to this adaptation is the evolution of genes that encode amidohydrolase (amidase) and other synthesis proteins that deaminate acrylamide to acrylic acid and ammonium (Shanker et al., 1990; Nawaz et al., 1994; Nawaz et al., 1998; Buranasilp and Charoenpanich, 2011; Thanyacharoen et al., 2012). With this, acrylic acid can be changed to propionate and subsequently succinyl CoA (Stams et al., 1993; Ansede et al., 1999; Wampler and Ensign, 2005) to generate energy. Potentially harmful ammonium is detoxified and MscL protein is released from the cell.
Classical proteomic approaches mostly provide information on relative amounts of protein species and only rarely information on activity. Precise functional understanding of the biological systems requires metabolomics and interaction studies. In conclusion, this paper provides important missing information on bacterial responses to acrylamide. Further transcriptional and translational studies would lead to the total resolution of the acrylamide degradation.
ACKNOWLEDGEMENTS
This work was made possible by funding support from the Agricultural Research Development Agency (Public Organization) to JC. This work is linked with the Asian Core Program (ACP) on capacity building and development of microbial potential and fermentation technology towards new era. We thank F. W. H. Beamish for editorial assistance.
REFERENCES
Ajouz, B., C. Berrier, A. Garrigues, M. Besnard, and A. Ghazi. 1998. Release of thioredoxin via the mechanosensitive channel MscL during osmotic downshock of Escherichia coli cells. Journal of Biological Chemistry 273: 26670-26674. 10.1074/jbc.273.41.26670
Ansede, J.H., P.J. Pellechia, and D.C. Yoch. 1999. Metabolism of acrylate to beta-hydroxypropionate and its role in dimethylsulfoniopropionate lyase induction by a salt marsh sediment bacterium, Alcaligenes faecalis M3A. Applied and Environmental Microbiology 65: 5075-5081.
Barbosa, T.M., and S.T. Levy. 2000. Differential expression of over 60 chromosomal genes in Escherichia coli by constitutive expression of Mar A. Journal of Bacteriology 182: 3467-3474. 10.1128/JB.182.12.3467-3474.2000
Berrier, C., A. Garrigues, G. Richarme, and A. Ghazi. 2000. Elongation factor tu and dnak are transferred from the cytoplasm to the periplasm of Escherichia coli during osmotic downshock presumably via the mechanosensitive channel MscL. Journal of Bacteriology 182: 248-251.
Bradford, M.M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing, the principle of protein-dye binding. Analytical Biochemistry 72: 248-254.
Buranasilp, K., and J. Charoenpanich. 2011. Biodegradation of acrylamide by Enterobacter aerogenes isolated from wastewater in Thailand. Journal of Environmental Sciences 23(3): 396-403. 10.1016/S1001-0742(10)60422-6
Busenlehner, L.S., M.A. Pennella, and D.P. Giedroc. 2003. The SmtB/ArsR family of metalloregulatory transcriptional repressors: structural insights into prokaryotic metal resistance. FEMS Microbiology Reviews 27: 131-143. 10.1016/S0168-6445(03)00054-8
Cha, M., and Chambliss, G.H. 2011. Characterization of acrylamidase isolated from a newly isolated acrylamide-utilizing bacterium, Rastonia eutropha AUM-01. Current Microbiology. 62 (2): 671-678. 10.1007/S00284-010-9761-8
Cherry, A.B., A.F. Gabaccia, and H.W. Senn. 1956. The assimilation behavior of certain toxic organic compounds in natural waters. Sewage Industrial Wastes 28: 1137-1146.
Choe, L.H., and K.H. Lee. 2000. A comparison of three commercially available isoelectric focusing units for proteome analysis: The multiphor, the IPGphor and the protean IEF cell. Electrophoresis 21: 993-1000. 10.1002.(SiCi)1522-2683 (2000301)21:5<993::AID-ELPS993>3.0.CO; 2-9
Croll, B.T., G.H. Arkell, and R.P.J. Hodge: Residues of acrylamide in water. Water Res., 8, 989-993 (1974).
Fayet, O., T. Ziegelhoffer, and C. Georgopoulos. 1989. The groES and groEL heat shock gene products of Escherichia coli are essential for bacterial growth at all temperatures. Journal of Bacteriology 171: 1379-1385.
Fischer, H.M., M. Babst, T. Kaspar, G. Acuna, F. Arigoni, and H. Hennecke. 1993. One member of a groESL-like chaperonin multigene family in Bradyrhizobium japonicum is co-regulated with symbiotic nitrogen fixation genes. EMBO Journal 12 (7): 2901-2912.
Hirrlinger, B., A. Stolz, and H.J. Knackmuss. 1996. Purification and properties of an amidase from Rhodococcus erythropolis MP50 which enantioselectively hydrolyzes 2-arylpropionamides. Journal of Bacteriology 178: 3501-3507.
IARC. 1994. IARC Monographs on the evaluation of carcinogenic risks to humans. 60: 389.
Kimbara, K., T. Hashimoto, M. Fukuda, T. Koana, M. Takagi, M. Oishi, and K. Yano. 1989. Cloning and sequencing of two tandem genes involved in degradation of 2,3-dihydroxybiphenyl to benzoic acid in the polychlorinated biphenyl-degrading soil bacterium Pseudomonas sp. strain KKS102. Journal of Bacteriology 171: 2740-2747.
Klose, J. 1975. Protein mapping by combined isoelectric focusing and electrophoresis of mouse tissues: A novel approach to testing for induced point mutation in mammals. Humangenetik 26: 231-243.
Koebnik, R., K.P. Loeher, and P. Van Gelder. 2000. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Molecular Microbiology 37: 239-253.
Laemmli, U.K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680-685. 10.1038/227680a0
Nagasawa, T., and H. Yamada. 1989. Microbial transformation of nitriles. Trends in Biotechnology 7: 153-158. 10.1016/1067-7799(89)90026-7
Nawaz, M.S., S.M. Billedeau, and C.E. Cerniglia. 1998. Influence of selected physical parameters on the biodegradation of acrylamide by immobilized cells of Rhodococcus sp. Biodegradation 9: 381-387. 10.1023/A:1008383710019
Nawaz, M.S., W. Franklin, and C.E. Cerniglia. 1993. Degradation of acrylamide by immobilized cells of a Pseudomonas sp. and Xanthomonas maltophilia. Canadian Journal of Microbiology 39: 207-212.
Nawaz, M.S., A.A. Khan, J.E. Seng, J.E. Leakey, P.H. Siitonen, and C.E. Cerniglia. 1994. Purification and characterization of an amidase from an acrylamidedegrading Rhodococcus sp. Applied and Environmental Microbiology 60: 3343-3348.
Nikaido, H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiology and Molecular Biology Reviews 67: 593-656. 10.1128/mmbr.67.4.593-656.2003
O’ Farrell, P.H. 1975. High resolution two-dimensional electrophoresis of proteins. Journal of Biological Chemistry 250: 4007-4021.
Payne, J.W. 1980. Transport and utilization of peptides by bacteria. In: Microorganisms and Nitrogen Sources (Eds.: J.W. Payne). Wiley, Chichester & London. pp. 211-256.
Payne, J.W., and M.W. Smith. 1994. Peptide transport by microorganisms. Advances in Microbiology and Physiology 36: 1-80.
Pomposiello, P.J., M.H.J. Bennik, and B.Demple. 2001. Genome-wide transcriptional profiling of the Escherichia coli response to superoxide stress and sodium salicylate. Journal of Bacteriology 183: 3890-3902. 10.1128/JB.183.13.3890-3902.2001
Prabu, C.S., and A.J. Thatheyus. 2007. Biodegradation of acrylamide employing free and immobilized cells of Pseudomonas aeruginosa. International Biodeteriation and Biodegradation 60: 69-73. 10.1016/j.ibiod.2006.11.007
Prasad, D.Y. 1982. Polyacrylamide as a coagulant aid in water treatment. Chem Age India 34: 387-391.
Segerbäck, D., C.J. Calleman, J.L. Schroeder, L.G. Costa, and E.M. Faustman. 1995. Formation of N-7-(2-carbamoyl-2-hydroxyethyl) guanine in DNA of the mouse and the rat following intraperitoneal administration of [14C] acrylamide. Carcinogenesis 16: 1161-1165.
Shanker, R., C. Ramakrishna, and P.K. Seth. 1990. Microbial degradation of acrylamide monomer. Archives of Microbiology 154: 192-198. 10.1007/BF00423332
Shukor, M.Y., N. Gusmanizar, N.A. Azmi, M. Hamid, J. Ramli, N.A. Shamaan, and M.A. Syed. 2009a. Isolation and characterization of an acrylamidedegrading Bacillus cereus. Journal of Environmental Biology 30: 57-64.
Shukor, M.Y., N. Gusmanizar, J. Ramli, N.A. Shamaan, W.P. MacCormack, and M.A. Syed. 2009b. Isolation and characterization of an acrylamide-degrading Antarctic bacterium. Journal of Environmental Biology 30: 107-112.
Stams, A.J.M., J.B. Van Dijk, C. Dijkema, and C.M. Plugge. 1993. Growth of syntrophic propionate-oxidizing bacteria with fumarate in the absence of methanogenic bacteria. Applied and Environmental Microbiology 59(4): 1114-1119.
Thanyacharoen, U., A. Tani, and J. Charoenpanich. 2012. Isolation and characterization of Kluyvera georgiana strain with the potential for acrylamide biodegradation. Journal of Environmental Sciences and Health, Part A 47(11): 1491-1499. 10.1080/10934529.2012.680312
Thiery, A., M. Maestracci, A. Arnaud, P. Galzy, and M. Nicolas. 1986. Purification and properties of an acylamide amidohydrolase (EC. 3.5.1.4) with a wide activity spectrum from Brevibacterium sp. R 312. Journal of Basic Microbiology 26: 299-311. 10.1002/jobm.3620260512
Tilson, H.A., and P.A. Cabe. 1979. The effects of acrylamide given acutely or in repeated doses on fore- and hindlimb functions of rats. Toxicology and Applied Pharmacology 47: 253-260. 10.1016/0041-008X(79)90319-3
Wampler, D.A., and S.A. Ensign. 2005. Photoheterotrophic metabolism of acrylamide by a newly isolated strain of Rhodopseudomonas palustris. Applied and Environmental Microbiology 71: 5850-5857. 10.1128/AEM.71.10.5850-5857.2005
Wang, C.C., and C.M. Lee. 2001. Denitrification with acrylamide by pure culture of bacteria isolated from acrylonitrile-butadiene-styrene resin manufactured wastewater treatment system. Chemosphere 44: 1047-1053. 10.1016/S0045-6535(00)00503-8
Wang, C.C., C.M. Lee, and A.S. Wu. 2009. Acrylic acid removal from synthetic wastewater and industrial wastewater using Ralstonia solanacearum and Acidovorax avenae isolated from a wastewater treatment system manufactured with polyacrylonitrile fiber. Water Science and Technology 60: 3011-3016. 10.2166/WST.2009.710
Yamada, H., Y. Asano, T. Hino, and Y. Tani. 1979. Microbial utilisation of acrylonitrile. Journal of Fermentation Technology 57: 8-14.
Zabaznaya, E.V., S.V. Kozulin, and S.P. Voronin. 1998. Selection of strains transforming acrylonitrile and acrylamide into acrylic acid. Applied Biochemistry and Microbiology 34: 341-345.
Jittima Charoenpanich1,2,3* and Akio Tani4
1 Department of Biochemistry and Centre of Excellence for Innovation in Chemistry (PERCH-CIC), Faculty of Science, Burapha University, Bangsaen, Chonburi 20131, Thailand
2 Environmental Science Program, Faculty of Science, Burapha University, Bangsaen, Chonburi 20131, Thailand
3 Centre of Excellence on Environmental Health and Toxicology (CHE), Ministry of Education, Thailand
4 Institutes of Plant Science and Resources (ISPR), Okayama University, 2-20-1 Chuo, Kurashiki, Okayama 710-0046, Japan
*Corresponding author. E-mail: jittima@buu.ac.th
Total Article Views

