
Isolation of a Novel Molybdenum-reducing and Azo Dye Decolorizing Enterobacter sp. strain Aft-3 from Pakistan
Mohd Shukri Shakor, Khan Aftab, Masdor Norazliin, Halmi Mohd Izuan Effendi*, Abdullah Siti Rozaimah Sheikh and Suukor Mohd YunusPublished Date : 2019-08-23
DOI : 10.12982/cmujns.2016.0008
Journal Issues : Number2 ,May - August 2016
ABSTRACT
Removing pollutants, such as heavy metals and organic dyes, using bioremediation is the most economical approach over the long term, when other methods, such as physical or chemical, may not be effective or feasible. The heavy metal molybdenum is toxic to ruminants and spermatogenesis in many animals, while dyes, including Methyl Red, are mutagenic to animals. In this study, we report on the ability of a molybdenum-reducing bacterium isolated from contaminated soil to decolorize the dye Methyl Red. The bacterium reduced molybdate to Mo-blue optimally between pH 5.8 and 6.5 and at 37°C. Glucose was the best electron donor for supporting molybdate reduction, followed by lactose, sucrose, maltose, raffinose, mucate, d-mannose, cellobiose, d-adonitol, d-mannitol, melibiose, glycerol, l-rhamnose, d-sorbitol, and l-arabinose, in descending order. Other requirements included a phosphate concentration at 5 mM and a molybdate concentration between 20 and 25 mM. The absorption spectrum of the Mo-blue produced was similar to that produced by previous Mo-reducing bacterium, and closely resembled a reduced phosphomolybdate. Molybdenum reduction was inhibited by mercury (ii), silver (i) and copper (ii) at 2 mg/L by 74.6, 67.0 and 63.3%, respectively. Biochemical analysis resulted in a tentative identification of the bacterium as Enterobacter sp. strain Aft-3. The ability of this bacterium to detoxify molybdenum and decolorize azo dye makes this bacterium an important tool for bioremediation.
Keywords: Bioremediation, Molybdenum, Azo dye, Microplate format, Enterobacter sp., Characterization
INTRODUCTION
Using bioremediation to eliminate toxic heavy metals and organic pollutants, particularly when present at very low levels, is regarded as an economical approach
over the long term, where other methods, including physical or chemical, may not be effective. Molybdenum is an essential heavy metal in trace amount, but toxic to a variety of organisms at elevated levels (AhmadPanahi et al., 2014). It has many industrial uses, including as an alloying agent, a component of automobile engine anti-freeze, a component of corrosion resistant steel, and a lubricant in the form of molybdenum disulphide. The widespread industrial use of molybdenum has polluted water to levels in the hundreds of parts per million in Tokyo Bay, the Tyrol in Austria, and the Black Sea (Davis, 1991; Neunhäuserer et al., 2001). In addition, in the 1970s, it posed a health hazard as a significant pollutant in sewage sludge (Lahann, 1976). Molybdenum toxicity in inhibiting spermatogenesis and arresting embryogenesis in organisms, such as catfish and mice, at levels as low as several parts per million have been reported (Meeker et al., 2008; Bi et al., 2013; Zhai et al., 2013; Zhang et al., 2013). Furthermore, molybdenum is very toxic to ruminants, especially cows, at levels as low as several parts per million (Underwood, 1979; Kincaid, 1980).
Azo dyes, an organic pollutant, are often present as a co-pollutant alongside heavy metals. Azo dyes, including Methyl Red (Figure 1), are characterized by the presence of one or more azo groups. Azo dyes are the largest class of dyes, with numerous applications in textiles, printing, tannery, foodstuffs, hydrocarbon additives, and cosmetics. Nearly one million tons of basic and diazo direct dyes are being produced annually (Solís et al., 2012). This extensive usage has resulted in environmental pollution. A study of the toxicity of methyl red on the earthworm (Pheretima phosthuma) showed morphological symptoms were observed upon exposure of the dye. Mortality rate in terms of LD50 for Methyl Red was 218.77 mg l−l. Sodium dodecyl sulphate-Polyacrylamide Gel Electrophoresis was employed to study protein alteration in various parts of the earthworm. Variation was observed in the head, clitella, abdomen and various other organs of the earthworm (Gupte et al., 2008). Other studies have indicated that the dye showed some mutagenic properties (Wong and Yuen, 1996). Physiochemical methods in degrading azo dyes can generate toxic byproducts. Hence, microbiological degradation is an attractive alternative, as many microorganisms can degrade azo dyes into less toxic byproducts (Saratale et al., 2011). Microbiological degradation of azo dyes, including Methyl Red, generally proceeds via cleavage of the azo groups (–N=N–) by azo reductase. The dye becomes colorless when this bond is broken (Syed et al., 2009). Numerous microorganisms, including fungi, yeast, and bacteria, have been reported with the ability to degrade this dye (Solís et al., 2012).
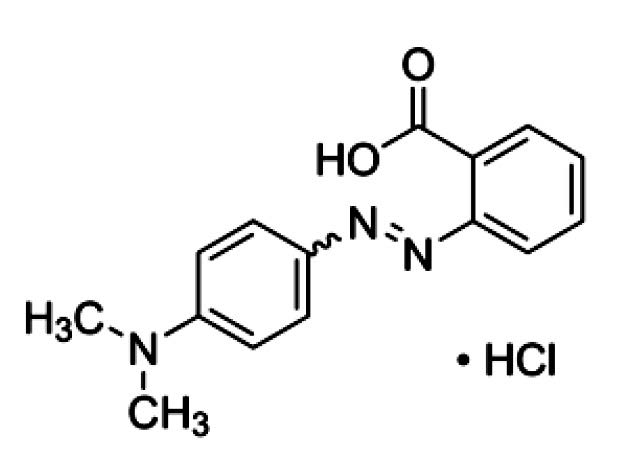
Figure 1. The structure of Methyl Red (Ng et al., 2014).
Since chromate reduction coupled with azo dye decolorization has been reported (Chaudhari et al., 2013), the aim of this study is to screen for the ability of a molybdenum (molybdate)-reducing bacterium to decolorize various dyes. Here we report molybdenum-reducing bacterium isolated from a contaminated soil with the novel capacity to decolorize the azo dye Methyl Red. The characteristics of this bacterium would make it suitable for future bioremediation applications involving both the heavy metal molybdenum and dye as an organic contaminant.
MATERIALS AND METHODS
Isolation of molybdenum-reducing bacterium
Soil samples were taken (5 cm deep from topsoil) from the grounds of a contaminated land in the pro-vince of Khyber, Pakhtunkhwa, Pakistan, in 2013. One gram of soil sample was suspended in sterile tap water. An aliquot of the soil suspension (0.1 mL) was pipetted and spread onto agar of low phosphate media (pH 7.0), and incubated for 24 hours at room temperature. The low phosphate media (LPM) was composed of: glucose (1%), (NH4)2.SO4 (0.3%), MgSO4.7H2O (0.05%), yeast extract (0.5%), NaCl (0.5%), Na2MoO4.2H2O (0.242 % or 10 mM), and Na2HPO4 (0.071% or 5 mM) (Yunus et al., 2009). The formation of blue colonies indicates molybdate reduction by molybdenum-reducing bacteria. The colony forming the strongest blue intensity was isolated and re-streaked on low phosphate media (LPM) to obtain pure culture. Molybdenum reduction in liquid media (at pH 7.0) was carried out in 100 mL of the above media in a 250 mL shake flask culture at room temperature for 48 hours on an orbital shaker set at 120 rpm with the same media above, but the phosphate concentration increased to 100 mM. The molybdenum blue (Mo-blue) absorption spectrum was studied by taking out 1.0 mL of the Mo-blue formed from the liquid culture above and then centrifuging at 10,000 x g for 10 minutes at room temperature. The supernatant was scanned at 400 to 900 nm using a UV-spectrophotometer (Shimadzu 1201). Low phosphate media was utilized as the baseline correction.
Morphological, physiological, and biochemical characterization of the isolated strain
Identification of the bacterium was carried out biochemically and phenotypically using standard methods, including colony shape, gram staining, size and color on nutrient agar plate, motility, oxidase (24 h), ONPG (beta-galactosidase), catalase production (24 h), ornithine decarboxylase (ODC), arginine dihydrolase (ADH), lysine decarboxylase (LDC), nitrates reduction, Methyl red, indole production, Voges-Proskauer (VP), hydrogen sulfide (H2S), acetate utilization, malonate utilization, citrate utilization (Simmons), esculin hydrolysis, tartrate (Jordans), gelatin hydrolysis, urea hydrolysis, deoxyribonuclease, lipase (corn oil), phenylalanine deaminase, gas production from glucose, and production of acids from various sugars, according to Bergey’s Manual of Determinative Bacteriology (Holt et al., 1994). Interpretation of the results was carried out via the ABIS online system (Costin and Ionut, 2015).
Preparation of resting cells for molybdenum reduction characterization
Characterization studies of molybdenum reduction to Mo-blue, such as the effects of pH, temperature, phosphate, and molybdate concentrations, were carried out statically using resting cells in a microplate or microtiter format, as previously developed (Shukor and Shukor, 2014). Cells from a 1 L overnight culture were grown in High Phosphate media (HPM) at room temperature on orbital shaker (150 rpm), with LPM and HPM differing only by the phosphate concentration, which was fixed at 100 mM for HPM. Cells were harvested by centrifugation at 15,000 x g for 10 minutes, and the pellet was washed several times to remove residual phosphate and resuspended in 20 mls of low phosphate media (LPM) minus glucose to an absorbance at 600 nm of approximately 1.00. In the low phosphate media, a concentration of 5 mM phosphate was optimal for all of the Mo-reducing bacteria isolated so far, as reported in the literature, and hence was used here. Higher concentrations were found to be strongly inhibitory to molybdate reduction (Campbell et al., 1985; Ghani et al., 1993; Shukor et al., 2007; Shukor et al., 2008a; Rahman et al., 2009; Shukor et al., 2009a; Yunus et al., 2009; Shukor et al., 2009b; Shukor et al., 2010a; Shukor et al., 2010b; Lim et al., 2012; Abo-Shakeer et al., 2013; Ahmad et al., 2013; Halmi et al., 2013; Othman et al., 2013; Khan et al., 2014). Then 180 μL was sterilely pipetted into each well of a sterile microplate. Sterile glucose (20 μL) from a stock solution was added to each well to initiate Mo-blue production. The effect of other carbon sources as electron donors for molybdenum reduction was studied by replacing glucose with lactose, sucrose, maltose, raffinose, mucate, d-mannose, cellobiose, d-adonitol, d-mannitol, melibiose, glycerol, l-rhamnose, d-sorbitol and l-arabinose, dulcitol, myo-inositol, and salicin.
A sterile sealing tape that allows gas exchange (Corning® microplate) was used to seal the microplate. The microplate was incubated at room temperature. At defined times, absorbance at 750 nm was read in a BioRad (Richmond, CA) Microtiter Plate reader (Model No. 680). The production of Mo-blue from the media in a microplate format was measured using the specific extinction coefficient of 11.69 mM.–1.cm–1 at 750 nm, as the maximum filter wavelength available for the microplate unit was 750 nm (Shukor et al., 2003).
Effect of heavy metals on molybdenum reduction
Ten heavy metals, namely chromium (vi), zinc (ii), mercury (ii), cadmium (ii), lead (ii), nickel (ii), copper (ii), arsenic (v), cobalt (ii), and silver (i) were prepared from commercial salts or from Atomic Absorption Spectrometry standard solutions from MERCK, and tested at the final concentration of 2 mg/L. The bacterium was incubated with heavy metals in the microplate format at various concentrations. The plate was incubated for 24 hours at 30°C. The amount of Mo-blue production was measured at 750 nm, as before.
Screening for bacterial decolorization of dyes
The ability of the bacterium to decolorize dyes was tested using the microplate format above, with the dyes added to the final concentration of 100 mg/L. All dyes were sourced from Sigma-Aldrich (St. Loius, U.S.A.). The dyes (with maximum wavelength in parentheses) were: Congo Red (C.I. 22120) (498 nm), Cresol Red (C.I. 1733-12-6) (570 nm), Crocein Orange G (C.I. 15970) (482 nm), Evans Blue (C.I. 23860) (594 nm), Fast Green FCF (C.I. 42053) (620 nm), Fuchsin Basic (C.I. 42510) (625 nm), Crystal Violet (C.I. 42555) (590 nm), Metanil Yellow (C.I. 13065) (414 nm), Methyl Green (C.I. 42590) (635 nm), Methyl Orange (C.I. 13025) (505 nm), Methyl Red (C.I. 13020) (493 nm), Methylene Blue (C.I. 52015) (590 nm), Naphthol Blue Black (C.I. 20470) (618 nm), Nigrosin (C.I. 50415) (570 nm), Orange G (C.I. 16230) (476 nm), Orange II sodium salt (C.I. 15510) (483 nm), Ponceau 2R (C.I. 16150) (388 nm), Ponceau S (C.I. 27195) (352 nm), Remazol Black B (C.I. 20505) (597 nm), Rhodamine B (C.I. 45170) (554 nm), Safranin O (C.I. 50240) (530 nm), Direct Blue 71 (C.I. 34140) (586 nm), Sudan Black B (C.I. 26150) (600 nm), Tartrazine (C.I. 19140) (427 nm), Toluidine Blue (C.I. 52040) (626 nm), and Trypan Blue (C.I. 23850) (607 nm).
The ingredients of the decolorizing media (% w/v) were as follows: glucose (1%), sodium lactate (1%), (NH4)2.SO4 (0.3%), NaNO3 (0.2%), MgSO4.7H2O (0.05%), yeast extract (0.05%), NaCl (0.5%), and Na2HPO4 (0.705% or 50 mM). Lactate and nitrate were added to enhance decolorization, as these chemical compounds have been found to accelerate decolorization of azo dyes in certain bacteria (Ng et al., 2014). The media was adjusted to pH 7.0. Some of the dyes change in color as the pH changes, so the phosphate concentration was increased to 50 mM at pH 7.0 to prevent this. Decolorization was monitored using three standard wavelengths – 405, 490, and 595 nm – to cover the maximum absorption values for specific dyes, as these wavelengths were available in the BioRad 680 microplate reader. In addition, the use of these preset wavelengths took into account that the maximum absorption spectrums of water soluble dyes are generally shallow and a ±20 nm difference from the maximum absorption wavelength does not dramatically reduce absorbance values. The difference of absorbance values from the initial measurements were subtracted from the final measurements after an incubation period of 48 hours and a percentage decolorization was calculated.
Statistical analysis
Values are means ± SE. Data analyses were carried out using Graphpad Prism version 3.0 and Graphpad InStat version 3.05, available from www.graphpad.com. A Student’s t-test or a one-way analysis of variance with post hoc analysis by Tukey’s test was employed to compare between groups. P < 0.05 was considered statistically significant.
RESULTS
Identification of molybdenum-reducing bacterium
The bacterium was a short rod-shaped, motile, Gram-negative and facultative anaerobe bacterium. The colonies were between 1 to 3 mm in diameter, shiny, cream white, smooth, and circular in shape. The bacterium was identified by comparing the results of cultural, morphological, and various biochemical tests (Table 1) to the Bergey’s Manual of Determinative Bacteriology (Holt et al., 1994) and using the ABIS online software (Costin and Ionut, 2015). The software gave three suggestions for the bacterial identity, with the highest homology (87%) and accuracy at 100% as Enterobacter aerogenes. We are currently sequencing the 16s rRNA gene from this bacterium for its molecular identification to more definitively identify the species. However, for the purposes of this research, the bacterium is tentatively identified as Enterobacter sp. strain Aft-3.
Table 1. Biochemical tests for Enterobacter sp. strain Aft-3.
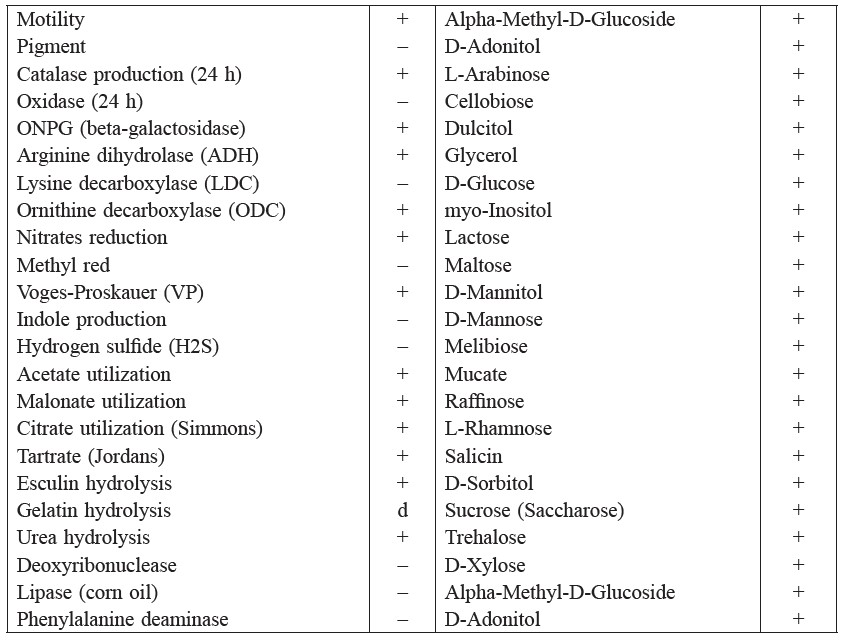
Note: + positive result, − negative result, d indeterminate result.
Molybdenum absorbance spectrum
The absorption spectrum of Mo-blue produced by Enterobacter sp. strain Aft-3 exhibited a shoulder at approximately 700 nm and a maximum peak near the infra-red region of between 860 and 870 nm, with a median at 865 nm (Figure 2). The identity of the Mo-blue is not easily ascertained, as it is complex in structure and has many species (Shukor et al., 2007). Briefly, Mo-blue is a reduced product of two types of molybdenum complexes – isopolymolybdate and heteropolymolybdate. It has been suggested by Campbell et al. (1985) that the Mo-blue observed in the reduction of molybdenum by E. coli K12 is a reduced form of phosphomolybdate, although they did not provide a plausible mechanism.
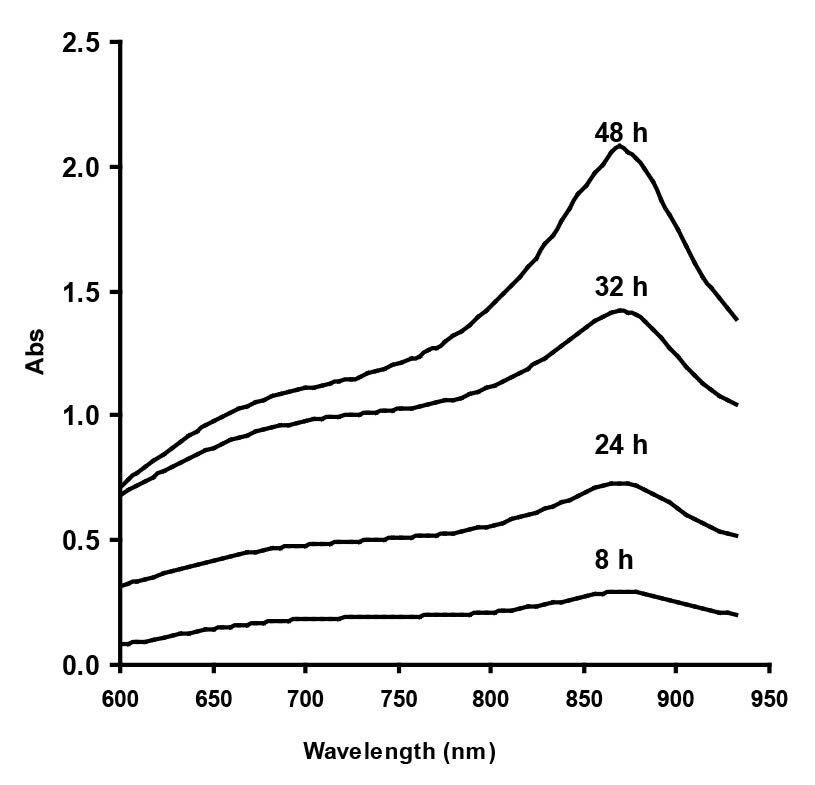
Figure 2. Scanning absorption spectrum of Mo-blue from Enterobacter sp. strain Aft-3 at different time intervals.
Effect of pH and temperature on molybdate reduction
Enterobacter sp. strain Aft-3 was incubated at different pH, ranging from 5.5 to 8.0, using Bis-Tris and Tris.Cl buffers (20 mM). Analysis by ANOVA showed that the optimum pH for reduction was between 6.0 and 6.5. Inhibition of reduction was dramatic at pH lower than 5 (Figure 3). The effect of temperature (Figure 4) was observed over a wide range (20 to 60°C), with an optimum temperature range of 30°C to 37°C with no significant difference (p>0.05) among the values measured, as analysed using ANOVA. Temperatures lower than 30°C and higher than 37°C were strongly inhibitory to Mo-blue production from Enterobacter sp. strain Aft-3.
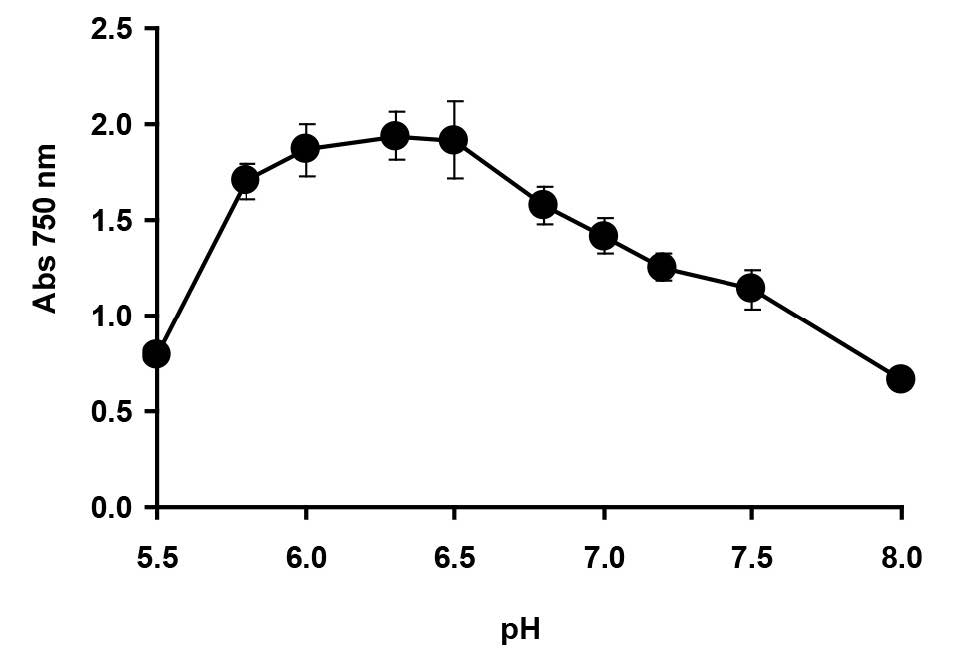
Figure 3. Effect of pH on molybdenum reduction by Enterobacter sp. strain Aft-3. Resting cells of the bacterium were incubated in a microtiter plate under optimized conditions for 48 hours. Error bars represent mean ± standard deviation (n=3).
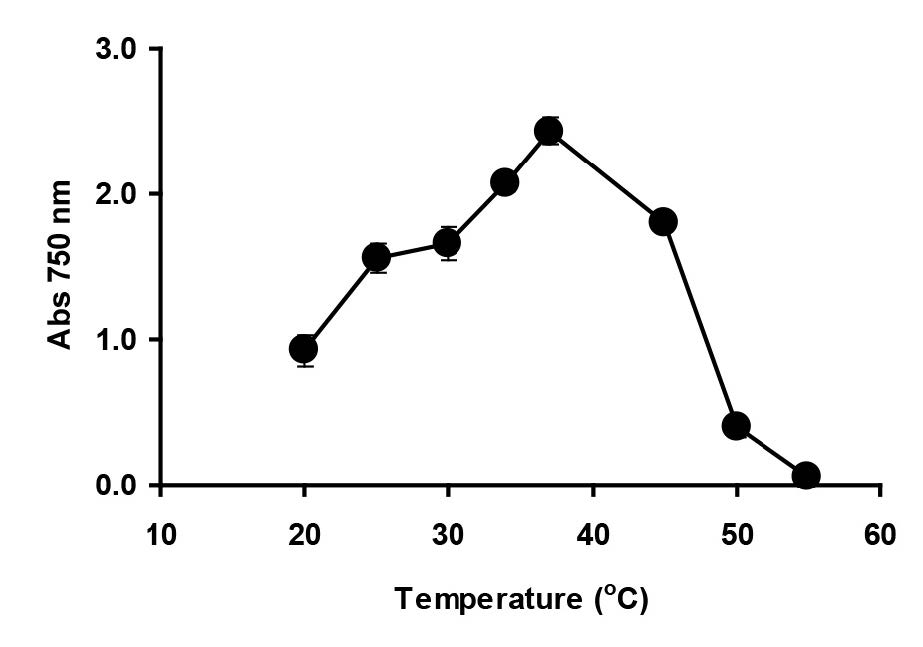
Figure 4. Effect of temperature on molybdenum reduction by Enterobacter sp. strain Aft-3. Resting cells of the bacterium were incubated in a microtiter plate under optimized conditions for 48 hours. Error bars represent mean ± standard deviation (n=3).
Effect of electron donor on molybdate reduction
Among the electron donors tested, glucose was the best for supporting molybdate reduction, followed by lactose, sucrose, maltose, raffinose, mucate, d-mannose, cellobiose, d-adonitol, d-mannitol, melibiose, glycerol, l-rhamnose, d-sorbitol, and l-arabinose, in descending order (Figure 5). Other carbon sources did not support molybdenum reduction.
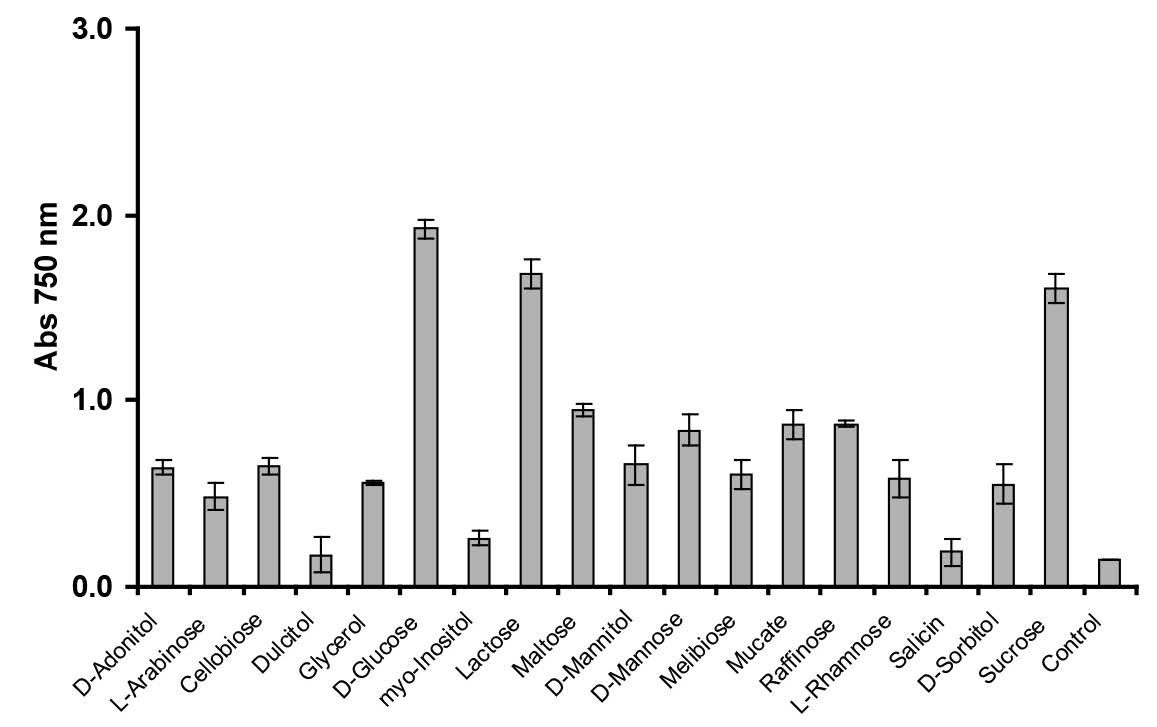
Figure 5. Effect of different electron donor sources (1% w/v) on molybdenum reduction. Enterobacter sp. strain Aft-3 was grown in low-phosphate media containing 10 mM molybdate and various electron donors. Resting cells of the bacterium were incubated in a microtiter plate under optimized conditions for 48 hours. Error bars represent mean ± standard deviation (n = 3).
Effect of phosphate and molybdate concentrations on molybdate reduction
The determination of phosphate and molybdate concentrations supporting optimal molybdenum reduction is important, because both anions have been shown to inhibit Mo-blue production in bacteria (Shukor et al., 2008a; Shukor et al., 2009a; Shukor et al., 2009b; Yunus et al., 2009; Shukor et al., 2010a; Shukor et al., 2010b; Lim et al., 2012; Ahmad et al., 2013; Othman et al., 2013; Shukor et al., 2014; Masdor et al., 2015). The optimum concentration of phosphate occurred at 5 mM; higher concentrations strongly inhibited reduction (Figure 6). High phosphate has been suggested to inhibit phosphomolybdate stability, as the complex requires acidic conditions and phosphate is a strong buffer. In addition, the phosphomolybdate complex itself is unstable in the presence of high phosphate through an unknown mechanism (Glenn and Crane, 1956).
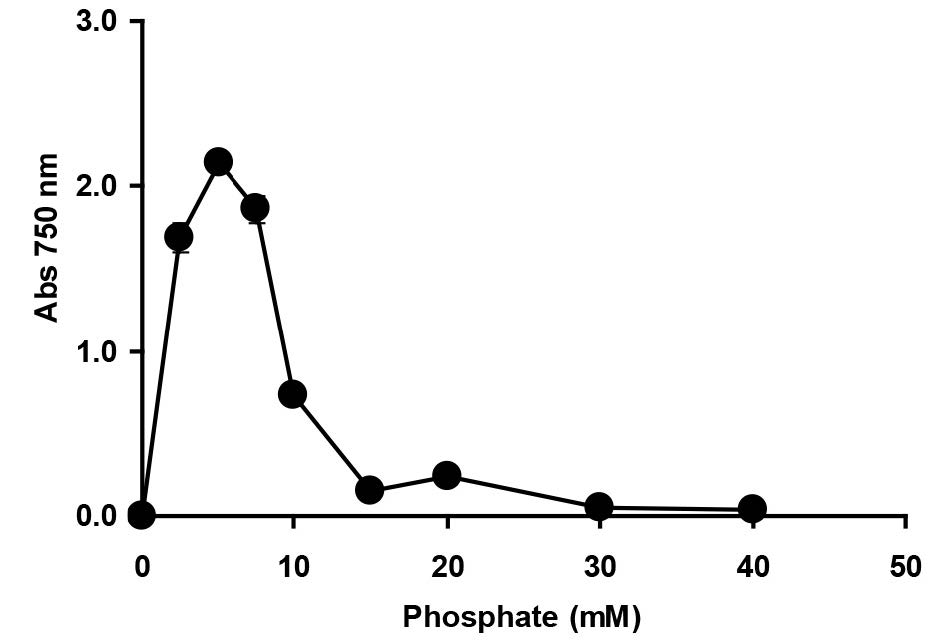
Figure 6. The effect of phosphate concentration on molybdenum reduction by Enterobacter sp. strain Aft-3. Resting cells of the bacterium were incubated in a microtiter plate under optimized conditions for 48 hours. Error bars represent mean ± standard deviation (n = 3).
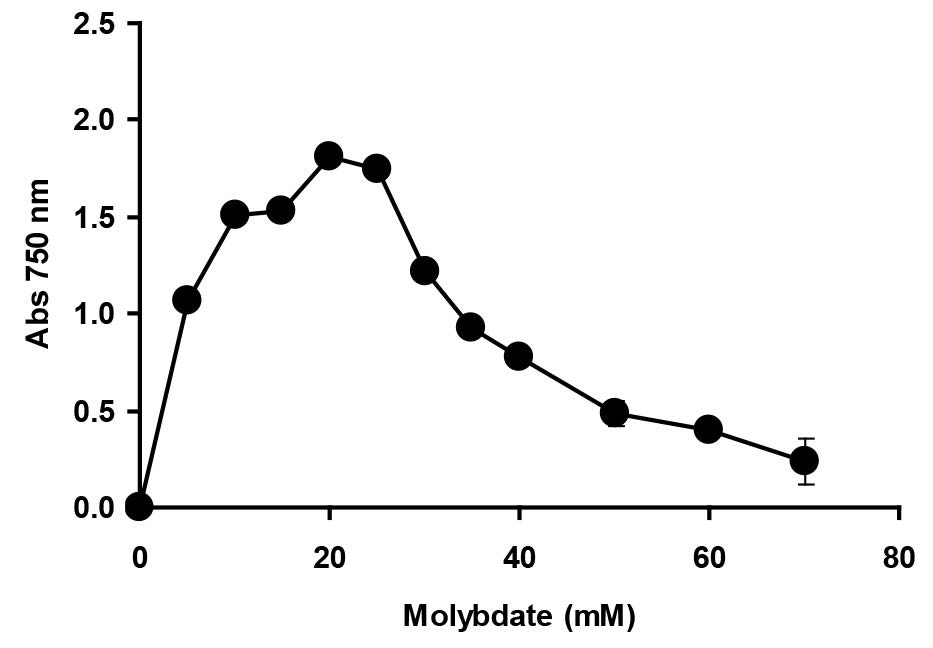
Figure 7. The effect of molybdate concentration on molybdenum reduction by Enterobacter sp. strain Aft-3. Resting cells of the bacterium were incubated in a microtiter plate under optimized conditions for 48 hours. Error bars represent mean ± standard deviation (n = 3).
Effect of heavy metals
Molybdenum reduction was inhibited by mercury (ii), silver (i), and copper (ii) at 2 mg/L by 74.6, 67.0 and 63.3%, respectively (Figure 8). The inhibition effects by other metal ions and heavy metals present a major problem for bioremediation. Therefore, it is important to screen and isolate bacteria for resistance to as many metals as possible.
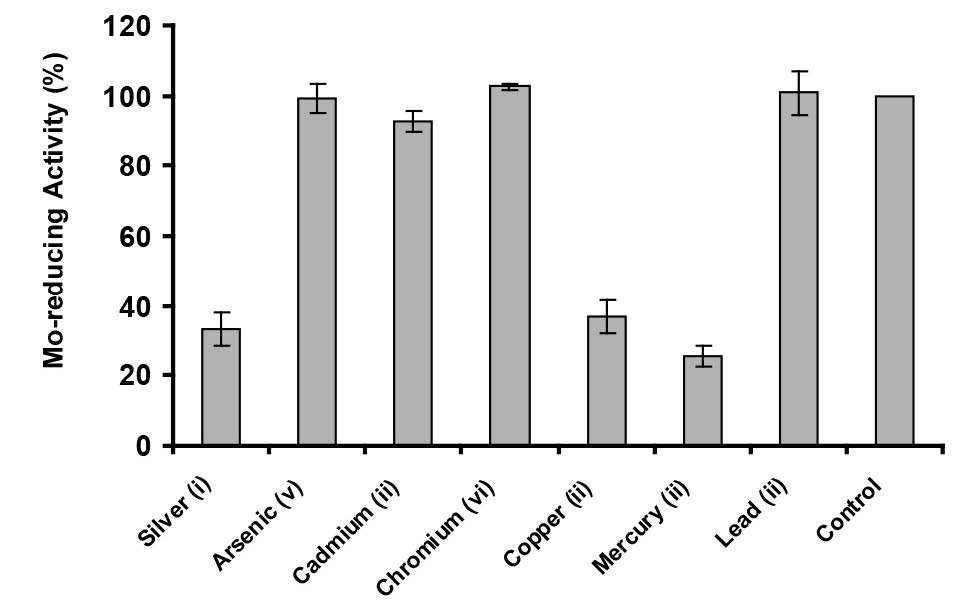
Figure 8. The effect of metals on Mo-blue production by Enterobacter sp. strain Aft-3. Resting cells of the bacterium were incubated in a microtiter plate under optimized conditions for 48 hours. Error bars represent mean ± standard deviation (n = 3).
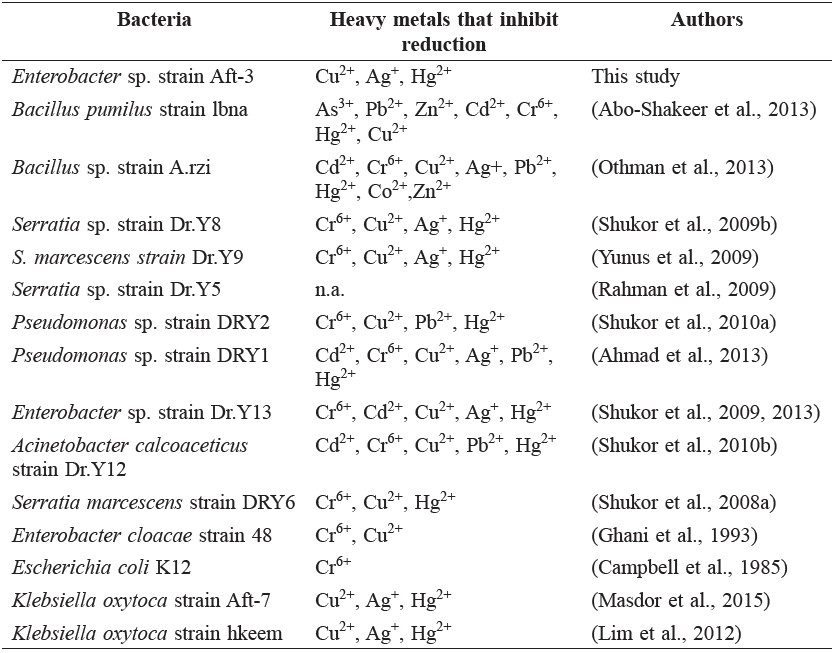
Table 2. Inhibition of Mo-reducing bacteria by heavy metals.
Azo dye-decolorizing ability of the molybdenum-reducing bacterium
We purposely used static growth conditions, which was easily achieved in a microplate environment where the oxygen concentration (0~10% environmentally oxygen, EO) was lower than aerobic conditions (~20% environmental oxygen, EO), as most bioremediation would have to be carried out in aquatic bodies or soils where the EO level is less than ~20% EO (Haley et al., 2012) and other electron acceptors, such as nitrates, would come into play. Nearly all of the molybdenum-reducing bacteria isolated so far reduced molybdenum into Mo-blue under static conditions.
The bacterium was able to decolorize the dye Methyl Red (Figure 9).
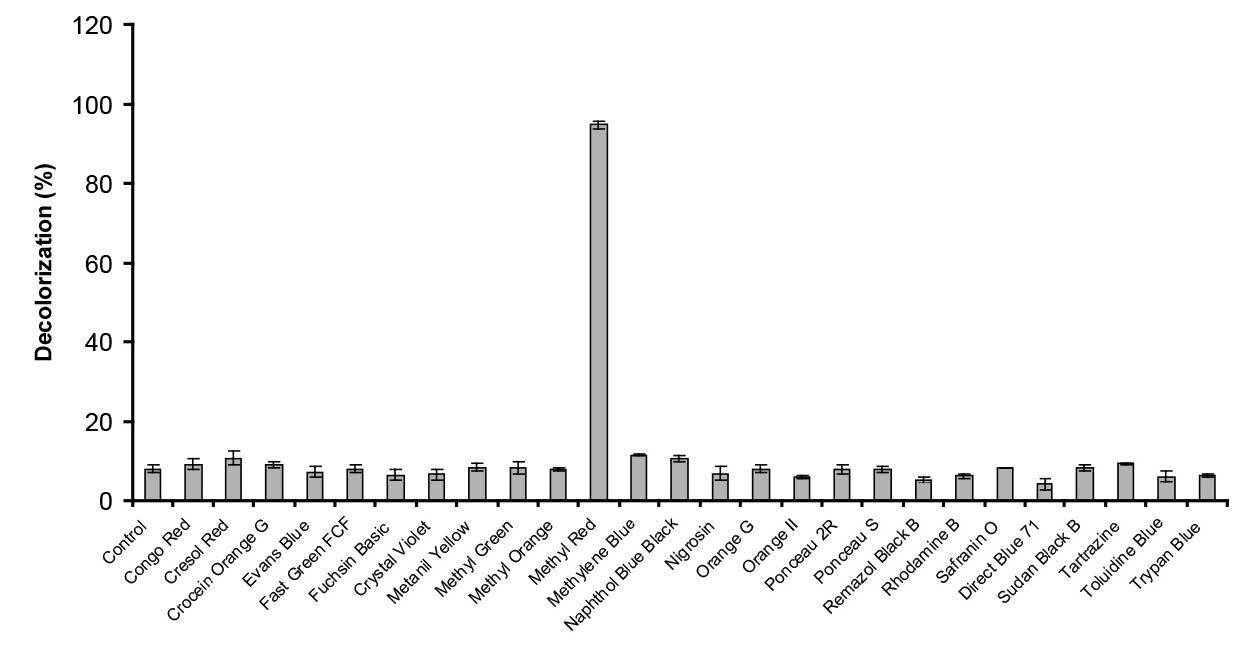
Figure 9. Decolorization of various dyes by Enterobacter sp. strain Aft-3. Resting cells of the bacterium were incubated in a microtiter plate under optimized conditions for 48 hours. Error bars represent mean ± standard deviation (n = 3).
DISCUSSION
The phenomenon of molybdenum reduction to molybdenum blue when sodium molybdate at concentrations of above 1 mM is supplemented to bacterial growth media was first reported more than a century ago, in 1896, in E. coli (Capaldi and Proskauer, 1896). Sporadic reporting of this phenomenon occurred in 1939 (Jan, 1939), 1985 (Campbell et al., 1985), and 1993 (Ghani et al., 1993). The potential of using molybdenum reduction for bioremediation of molybdenum was first mentioned by Ghani et al. (Ghani et al., 1993). Since then, numerous Mo-reducing bacteria have been isolated (Campbell et al., 1985; Ghani et al., 1993; Shukor et al., 2008a; Rahman et al., 2009; Shukor et al., 2009a; Shukor et al., 2009b; Shukor et al., 2010a; Shukor et al., 2010b; Lim et al., 2012; Abo-Shakeer et al., 2013; Halmi et al., 2013; Othman et al., 2013; Khan et al., 2014; Shukor et al., 2014), including two bacteria that can degrade SDS (Halmi et al., 2013; Masdor et al., 2015), as well as a psychrotolerant Mo-reducing bacterium isolated from Antarctica (Ahmad et al., 2013). A rapid and simple high throughput method involving the use of a microplate was utilized to speed-up characterization to obtain more data than the normal shake-flask approach (Shukor and Shukor, 2014). The use of resting cells under static conditions to characterize molybdenum reduction in bacterium was initiated by Ghani et al. (1993). Resting cells have been used to study heavy metals reduction, such as in chromate (Llovera et al., 1993) and vanadate (Carpentier et al., 2005) reductions.
The molybdenum blue produced by bacterial reduction of molybdenum has been studied in detail, due to the complexity of this compound’s structure, which exists in numerous forms or species, as well as its ability to change from one species to another, if the pH of the environment changes (Sims, 1961). Molybdenum blue can be produced from the reduction of isopolymolybdate or heteropolymolybdate to isopolymolybdenum blue or heteropolymolybdenum blue, respectively. The formation of isopoly Mo-blue from molybdate itself is not possible using biological-based reducing agents, as the conversion requires strong reducing agents under acidic conditions. The formation of heteropoly Mo-blue by biologically-based reducing agents, such as ascorbic acids or enzymatic reduction, is more plausible, as seen in the phosphate determination method using ascorbic acid. The Mo-blue produced from this method is a reduced phosphomolybdate, a heteropolymolybdate (Glenn and Crane, 1956). We hypothesize that microbial molybdate reduction in media containing molybdate and phosphate must proceed via the phosphomolybdate intermediate, and the conversion from molybdate to this structure occurs due to the reduction of pH during bacterial growth; in other words, the reduction of molybdenum to Mo-blue requires both chemical and biological processes. The absorption spectrum of the Mo-blue from this bacterium if it goes through this mechanism should show a spectrum closely resembling the phosphate determination method. To be exact, the spectrum observed showed a maximum absorption between 860 and 870 nm, and a shoulder at approximately 700 nm (Shukor et al., 2007). The Mo-blue spectrum from phosphate determination normally showed a maximum absorption around 880 to 890 nm, and a shoulder around 700 to 720 nm (Glenn and Crane, 1956; Sims, 1961). We have shown previously that the entire Mo-blue spectra from other bacteria obey this requirement (Shukor et al. 2007). In this study, the result from the absorption spectrum clearly implies a similar spectrum, and thus provides evidence for the hypothesis. Exact identification of the phosphomolybdate species must be carried out using nuclear magnetic resonance and electron spin resonance, due to the complex structure of the compound (Sims, 1961). However, spectrophotmetric characterization of heteropolymolybdate species via analyzing the scanning spectroscopic profile is a less cumbersome but accepted method (Glenn and Crane, 1956; Sims, 1961). Although the maximum absorption wavelength for Mo-blue was 865 nm, measuring at 750 nm, although approximately 30% lower, was enough for routine monitoring of Mo-blue production, as the intensity obtained was much higher than cellular absorption at 600-620 nm (Shukor and Shukor, 2014). Previous monitoring of Mo-blue production used several wavelengths, such as 710 nm (Ghani et al., 1993) and 820 nm (Campbell et al., 1985).
Temperature and pH play important roles in molybdenum reduction; since this process is enzyme mediated, both parameters affect protein folding and enzyme activity, causing the inhibition of molybdenum reduction. The optimum conditions would be an advantage for bioremediation in a tropical country, like Malaysia, which has an average yearly temperature ranging from 25 to 35°C (Shukor et al., 2008a). Therefore, Enterobacter sp. strain Aft-3 could be a candidate for soil bioremediation of molybdenum in the tropics. The majority of the reducers shows an optimal temperature of between 25 and 37°C (Shukor et al., 2008a; Rahman et al., 2009; Shukor et al., 2009a; Yunus et al., 2009; Shukor et al., 2010a; Shukor et al., 2010b; Lim et al., 2012; Abo-Shakeer et al., 2013; Halmi et al. 2013; Othman et al. 2013; Khan et al., 2014; Shukor et al., 2014; Masdor et al., 2015), as they are isolated from tropical soils; the only psychrotolerant reducer, isolated from Antartica, showed an optimal temperature supporting reduction of between 15 and 20°C (Ahmad et al., 2013).
The optimal pH range exhibited by Enterobacter sp. strain Aft-3 for supporting molybdenum reduction reflects the property of the bacterium as a neutrophile, which are able to grow between pH 5.5 and 8.0. An important observation regarding molybdenum reduction in bacteria is the optimal pH reduction is slightly acidic, with optimal pHs ranging from pH 5.0 to 7.0 (Campbell et al., 1985; Ghani et al., 1993; Shukor et al., 2008b; Rahman et al., 2009; Shukor et al., 2009a; Shukor et al., 2009b; Shukor et al., 2010a; Shukor et al., 2010b; Lim et al., 2012; Abo-Shakeer et al., 2013; Ahmad et al., 2013; Halmi et al., 2013; Othman et al., 2013; Khan et al., 2014; Shukor et al., 2014). It has been suggested previously that acidic pH plays an important role in the formation and stability of phosphomolybdate, before its reduction to Mo-blue. Thus, the optimal reduction occurs by balancing enzyme activity and substrate stability (Shukor et al., 2007).
Previous works by Shukor et al. demonstrated that the best carbon source for several Mo-reducing bacteria, such as Enterobacter cloacae strain 48 (Ghani et al., 1993), Serratia sp. strain Dr.Y5 (Rahman et al., 2009), S. marcescens strain Dr.Y9 (Yunus et al., 2009), and Serratia marcescens strain DRY6 (Shukor et al., 2008a), was sucrose. Other molybdenum reducers, such as Escherichia coli K12 (Campbell et al., 1985), Serratia sp. strain Dr.Y5 (Rahman et al., 2009), Pseudomonas sp. strain DRY2 (Shukor et al., 2010a), Pseudomonas sp. strain DRY1 (Ahmad et al., 2013), Enterobacter sp. strain Dr.Y13 (Shukor et al., 2009a), Acinetobacter calcoaceticus strain Dr.Y12 (Shukor et al., 2010b), Bacillus pumilus strain lbna (Abo-Shakeer et al., 2013), Klebsiella oxytoca strain AFT-7 (Masdor et al., 2015), and Bacillus sp. strain A.rzi (Othman et al., 2013), prefer glucose as the carbon source, while the Klebsiella oxytoca strain hkeem prefers fructose (Lim et al., 2012). In the presence of carbon sources in the media, the bacteria could produce electron donating substrates, NADH and NADPH, through metabolic pathways, such as glycolysis, Kreb’s cycle, and electron transport chain. Both NADH and NADPH serve as the electron donating substrates for molybdenum reducing-enzyme (Shukor et al., 2008b; Shukor et al., 2014).
All of the molybdenum-reducing bacterium isolated so far required phosphate concentrations not higher than 5 mM for optimal reduction (Campbell et al., 1985; Ghani et al., 1993; Shukor et al., 2008a; Rahman et al., 2009; Shukor et al., 2009a; Shukor et al., 2009b; Shukor et al., 2010a; Shukor et al., 2010b; Lim et al., 2012; Abo-Shakeer et al., 2013; Ahmad et al., 2013; Halmi et al., 2013; Othman et al., 2013; Khan et al., 2014; Shukor et al., 2014; Masdor et al., 2015). Studies on the effect of molybdenum concentration on molybdenum reduction showed that the newly isolated bacterium was able to reduce molybdenum as high as 60 mM, but with reduced Mo-blue production. The optimal reduction range was between 20 and 25 mM (Figure 7). Reduction at this high of a concentration into an insoluble form would allow the strain to reduce high concentrations of molybdenum pollution. The lowest optimal concentration of molybdenum reported was 15 mM in Pseudomonas sp strain Dr.Y2 (Shukor et al., 2010a), whilst the highest molybdenum required for optimal reduction was 80 mM in E. coli K12 (Campbell et al., 1985) and Klebsiella oxytoca strain hkeem (Lim et al., 2012). Other Mo-reducing bacteria, such as EC48 (Ghani et al., 1993), S. marcescens strain Dr.Y6 (Shukor et al., 2008a), S. marcescens. Dr.Y9 (Yunus et al., 2009), Pseudomonas sp. strain Dr.Y2 (Shukor et al., 2010a), Serratia sp. strain Dr.Y5 (Rahman et al., 2009), Enterobacter sp. strain Dr.Y13 (Shukor et al., 2009a), and Acinetobacter calcoaceticus (Shukor et al., 2010b), produced optimal Moblue at optimal molybdate concentrations of 50, 25, 55, 30, 30, 50, and 20 mM, respectively. In fact, the highest environmental concentration of molybdenum as a pollutant is around 2000 ppm, or about 20 mM (Runnells et al., 1976).
As described previously (Shukor et al., 2002), mercury is a physiological inhibitor to molybdate reduction. A summary of the type of heavy metals that inhibited Mo-reducing bacteria showed that almost all of the reducers were inhibited by toxic heavy metals (Table 2). Heavy metals, such as mercury, cadmium, silver, and copper, usually target the sulfhydryl group of enzymes (Llovera et al., 1993). Chromate is known to inhibit certain enzymes, such as glucose oxidase (Zeng et al., 2004) and enzymes of nitrogen metabolism in plants (Sangwan et al., 2014). Binding of heavy metals inactivated the metal-reducing capability of the enzyme(s) responsible for the reduction.
Azo dyes are strongly resistant to biodegradation under normal conditions, but the azo bond is vulnerable to reductive cleavage. Bacterial species that have been reported to be able to degrade this dye are Vibrio logei, Pseudomonas nitroreducens, Citrobacter sp., Sphingomonas paucimobilis, Lactobacillus acidophilus, Lactobacillus fermentum, Staphylococcus saprophyticus, Pseudomonas luteola, Bacillus sp. and Pseudomonas stutzeri, Enterobacter agglomerans, Aeromonas hydrophila, Acetobacter liquefaciens, Klebsiella pneumoniae, Bacillus subtilis, Enterococcus faecalis, P. aeruginosa, and Pseudomonas fluorescens (Saratale et al., 2011; Solís et al., 2012).
CONCLUSION
A local isolate of Mo-reducing bacterium with the novel ability to decolorize azo dye has been isolated. This is the first report of a molybdenum reducing bacterium with the ability to decolorize the dye Methyl Red. The bacterium reduces molybdate to Mo-blue optimally between pH 5.8 and 6.5 and at 37°C. Glucose was the best electron donor for supporting molybdate reduction, followed by lactose, sucrose, maltose, raffinose, mucate, d-mannose, cellobiose, d-adonitol, d-mannitol, melibiose, glycerol, l-rhamnose, d-sorbitol, and l-arabinose, in descending order. Other requirements included a phosphate concentration of 5 mM and a molybdate concentration between 20 and 25 mM. The absorption spectrum of the Mo-blue produced was similar to previous Mo-reducing bacterium, and closely resembled a reduced phosphomolybdate. Molybdenum reduction was inhibited by mercury (ii), silver (i), and copper (ii) at 2 mg/L by 74.6, 67.0 and 63.3%, respectively. The ability of this bacterium to detoxify multiple toxicants is a sought after property, and this makes the bacterium an important tool for bioremediation. Currently, work is underway to purify the molybdenum-reducing enzyme from this bacterium and to characterize decolorization studies in more detail.
ACKNOWLEDGMENTS
This project was supported by funds from Snoc International Sdn Bhd.
REFERENCES
Abo-Shakeer, L.K.A., S.A. Ahmad, M.Y. Shukor, N.A. Shamaan, and M.A. Syed. 2013. Isolation and Characterization of a Molybdenum-Reducing Bacillus Pumilus Strain Lbna. Journal of Environmental Microbiology and Toxicology. 1(1): 9-14.
AhmadPanahi, H., M. Hosseinzadeh, H. Adinehlo, E. Moniri, and M. Manoochehri. 2014. Removal of Molybdenum from Environmental Sample by Adsorption Using Modified Aniline-Formaldehyde with Salicylic Acid. World Applied Sciences Journal. 30(12): 1892-1898.
Ahmad, S.A., M.Y. Shukor, N.A. Shamaan, W.P. Mac Cormack, and M.A. Syed. 2013. Molybdate Reduction to Molybdenum Blue by an Antarctic Bacterium. BioMed Research International 2013. doi:10.1155/2013/871941.
Bi, C. M., Y. L. Zhang, F. J. Liu, T. Z. Zhou, Z. J. Yang, S. Y. Gao, S. D. Wang, et al. 2013. The Effect of Molybdenum on the in Vitro Development of Mouse Preimplantation Embryos. Systems Biology in Reproductive Medicine. 59(2): 69-73.
Campbell, A.M., A. Del Campillo-Campbell, and D.B. Villaret. 1985. Molybdate Reduction by Escherichia Coli K-12 and Its Chl Mutants. Proceedings of the National Academy of Sciences of the United States of America. 82(1): 227-231.
Capaldi, A., and B. Proskauer. 1896. Beiträge Zur Kenntniss Der Säurebildung Bei Typhus-Bacillen Und Bacterium Coli-Eine Differential-Diagnostische Studie. Zeitschrift Für Hygiene Und Infectionskrankheiten. 23(3): 452-474.
Carpentier, W., L. De Smet, J. Van Beeumen, and A. Brigé. 2005. Respiration and Growth of Shewanella Oneidensis MR-1 Using Vanadate as the Sole Electron Acceptor. Journal of Bacteriology. 187(10): 3293-3301.
Chaudhari, A.U., S.R. Tapase, V.L. Markad, and K.M. Kodam. 2013. Simultaneous Decolorization of Reactive Orange M2R Dye and Reduction of Chromate by Lysinibacillus Sp. KMK-A. Journal of Hazardous Materials. 262: 580-588.
Costin, S., and S. Ionut. 2015. ABIS Online-Bacterial Identification Software, http://www.tgw1916.net/bacteria_logare.html, Database Version: Bacillus 022012-2.10, Accessed on Mar 2015.
Davis, G.K. 1991. Molybdenum. In Metals and Their Compounds in the Environment, Occurrence, Analysis and Biological Relevance, edited by E. Merian. 1089-1100. VCH Weinheim, New York.
Ghani, B., M. Takai, N.Z. Hisham, N. Kishimoto, A.K.M. Ismail, T. Tano, and T. Sugio. 1993. Isolation and Characterization of a Mo6+-Reducing Bacterium. Applied and Environmental Microbiology. 59(4): 1176-1180.
Glenn, J.L., and F.L. Crane. 1956. Studies on Metalloflavoproteins. V. The Action of Silicomolybdate in the Reduction of Cytochrome c by Aldehyde Oxidase. Biochimica et Biophysica Acta. 22(1): 111-115.
Haley, C.L., J.A. Colmer-Hamood, and A.N. Hamood. 2012. Characterization of Biofilm-like Structures Formed by Pseudomonas Aeruginosa in a Synthetic Mucus Medium. BMC Microbiology 12.
Halmi, M.I.E., S.W. Zuhainis, M.T. Yusof, N.A. Shaharuddin, W. Helmi, Y. Shukor, M.A. Syed, and S.A. Ahmad. 2013. Hexavalent Molybdenum Reduction to Mo-Blue by a Sodium-Dodecyl-Sulfate-Degrading Klebsiella Oxytoca Strain dry14. BioMed Research International 2013 (Article ID 384541): 8 pages.
Holt, J.G., N.R. Krieg, P.H.A. Sneath, J.T. Staley, and S.T. Williams. 1994. Bergey’s Manual of Determinative Bacteriology. 9th ed. Lippincott Williams & Wilkins.
Jan, A. 1939. La Reduction Biologique Du Molybdate D’ammonium Par Les Bactéries Du Genre Serratia (The Biological Reduction of Ammonium Molybdate by the Bacteria of the Serratia Kind). Bulletin Des Sciences Pharmacologiques. 46: 336-339.
Khan, A., M.I.E. Halmi, and M.Y. Shukor. 2014. Isolation of Mo-Reducing Bacterium in Soils from Pakistan. Journal of Environmental Microbiology and Toxicology. 2(1): 38-41.
Kincaid, R. L. 1980. Toxicity of Ammonium Molybdate Added to Drinking Water of Calves. Journal of Dairy Science. 63(4): 608-610.
Lahann, R. W. 1976. Molybdenum Hazard in Land Disposal of Sewage Sludge. Water, Air, and Soil Pollution. 6(1): 3-8.
Lim, H.K., M.A. Syed, and M.Y. Shukor. 2012. Reduction of Molybdate to Molybdenum Blue by Klebsiella Sp. Strain Hkeem. Journal of Basic Microbiology. 52(3): 296-305.
Llovera, S., R. Bonet, M. D. Simon-Pujol, and F. Congregado. 1993. Chromate Reduction by Resting Cells of Agrobacterium Radiobacter EPS-916. Applied and Environmental Microbiology. 59(10): 3516-3518.
Masdor, N., M.S. Abd Shukor, A. Khan, M.I.E. Bin Halmi, S.R.S. Abdullah, N.A. Shamaan, and M.Y. Shukor. 2015. Isolation and Characterization of a Molybdenum-Reducing and SDS-Degrading Klebsiella Oxytoca Strain Aft-7 and Its Bioremediation Application in the Environment. Biodiversitas. 16(2): 238-246.
Meeker, J. D., M. G. Rossano, B. Protas, M. P. Diamond, E. Puscheck, D. Daly, N. Paneth, and J. J. Wirth. 2008. Cadmium, Lead, and Other Metals in Relation to Semen Quality: Human Evidence for Molybdenum as a Male Reproductive Toxicant. Environmental Health Perspectives. 116(11): 1473-1479.
Neunhäuserer, C., M. Berreck, and H. Insam. 2001. Remediation of Soils Contaminated with Molybdenum Using Soil Amendments and Phytoremediation. Water, Air, and Soil Pollution. 128(1-2): 85-96.
Ng, I.-S., T. Chen, R. Lin, X. Zhang, C. Ni, and D. Sun. 2014. Decolorization of Textile Azo Dye and Congo Red by an Isolated Strain of the Dissimilatory Manganese-Reducing Bacterium Shewanella Xiamenensis BC01. Applied Microbiology and Biotechnology. 98(5): 2297-2308.
Othman, A.R., N.A. Bakar, M.I.E. Halmi, W.L.W. Johari, S.A. Ahmad, H. Jirangon, M.A. Syed, and M.Y. Shukor. 2013. Kinetics of Molybdenum Reduction to Molybdenum Blue by Bacillus Sp. Strain A.rzi. BioMed Research International 2013. doi:10.1155/2013/371058.
Rahman, M.F.A., M.Y. Shukor, Z. Suhaili, S. Mustafa, N.A. Shamaan, and M.A. Syed. 2009. Reduction of Mo(VI) by the Bacterium Serratia Sp. Strain DRY5. Journal of Environmental Biology. 30(1): 65-72.
Runnells, D.D., D.S. Kaback, and E.M. Thurman. 1976. Geochemistry and Sampling of Molybdenum in Sediments, Soils, and Plants in Colorado. In Molybdenum in the Environment, edited by W.R. Chappel and K.K. Peterson. New York: Marcel and Dekker, Inc.
Sangwan, P., V. Kumar, and U. N. Joshi. 2014. Effect of chromium(VI) Toxicity on Enzymes of Nitrogen Metabolism in Clusterbean (Cyamopsis Tetragonoloba L.). Enzyme Research 2014: 784036.
Saratale, R. G., G. D. Saratale, J. S. Chang, and S. P. Govindwar. 2011. Bacterial Decolorization and Degradation of Azo Dyes: A Review. Journal of the Taiwan Institute of Chemical Engineers. 42(1): 138-157.
Shukor, M.S., and M.Y. Shukor. 2014. A Microplate Format for Characterizing the Growth of Molybdenum-Reducing Bacteria. Journal of Environmental Microbiology and Toxicology. 2(2): 42-44.
Shukor, M.Y., S.A. Ahmad, M.M.M. Nadzir, M.P. Abdullah, N.A. Shamaan, and M.A. Syed. 2010a. Molybdate Reduction by Pseudomonas Sp. Strain DRY2. Journal of Applied Microbiology. 108(6): 2050-2058.
Shukor, M.Y., S.H.M. Habib, M.F.A. Rahman, H. Jirangon, M.P.A. Abdullah, N.A. Shamaan, and M.A. Syed. 2008a. Hexavalent Molybdenum Reduction to Molybdenum Blue by S. Marcescens Strain Dr. Y6. Applied Biochemistry and Biotechnology. 149(1): 33-43.
Shukor, M.Y., M.I.E. Halmi, M.F.A. Rahman, N.A. Shamaan, and M.A. Syed. 2014. Molybdenum Reduction to Molybdenum Blue in Serratia Sp. Strain DRY5 Is Catalyzed by a Novel Molybdenum-Reducing Enzyme. BioMed Research International 2014. doi:10.1155/2014/853084.
Shukor, M.Y., C.H. Lee, I. Omar, M.I.A. Karim, M.A. Syed, and N.A. Shamaan. 2003. Isolation and Characterization of a Molybdenum-Reducing Enzyme in Enterobacter Cloacae Strain 48. Pertanika Journal of Science and Technology. 11(2): 261-272.
Shukor, M.Y., M.F.A. Rahman, N.A. Shamaan, C.H. Lee, M.I.A. Karim, and M.A. Syed. 2008b. An Improved Enzyme Assay for Molybdenum-Reducing Activity in Bacteria. Applied Biochemistry and Biotechnology. 144(3): 293-300.
Shukor, M.Y., M.F. Rahman, N.A. Shamaan, and M.S. Syed. 2009a. Reduction of Molybdate to Molybdenum Blue by Enterobacter Sp. Strain Dr.Y13. Journal of Basic Microbiology 49 (SUPPL. 1): S43-54.
Shukor, M.Y., M.F. Rahman, Z. Suhaili, N.A. Shamaan, and M.A. Syed. 2009b. Bacterial Reduction of Hexavalent Molybdenum to Molybdenum Blue. World Journal of Microbiology and Biotechnology. 25(7): 1225-1234.
Shukor M.Y., M.F. Rahman, Z. Suhaili, N.A. Shamaan, M.A. Syed. 2010b. Hexavalent Molybdenum Reduction to Mo-Blue by Acinetobacter Calcoaceticus. Folia Microbiologica. 55(2): 137-143.
Shukor, M.Y., M.A. Syed, C.H. Lee, M.I.A. Karim, and N.A. Shamaan. 2002. A Method to Distinguish between Chemical and Enzymatic Reduction of Molybdenum in Enterobacter Cloacae Strain 48. Malaysian Journal of Biochemistry 7: 71-72.
Shukor, Y., H. Adam, K. Ithnin, I. Yunus, N.A. Shamaan, and A. Syed. 2007. Molybdate Reduction to Molybdenum Blue in Microbe Proceeds via a Phosphomolybdate Intermediate. Journal of Biological Sciences. 7(8): 1448-1452.
Sims, R.P.A. 1961. Formation of Heteropoly Blue by Some Reduction Procedures Used in the Micro-Determination of Phosphorus. The Analyst 86 (1026): 584-590.
Solís, M., A. Solís, H.I. Pérez, N. Manjarrez, and M. Flores. 2012. Microbial Decolouration of Azo Dyes: A Review. Process Biochemistry. 47(12): 1723-1748.
Syed, M. A., H. K. Sim, A. Khalid, and M. Y. Shukor. 2009. A Simple Method to Screen for Azo-Dye-Degrading Bacteria. Journal of Environmental Biology. 30(1): 89-92.
Underwood, E. J. 1979. Environmental Sources of Heavy Metals and Their Toxicity to Man and Animals. InInternational Conference on Development in Land Methods of Wastewater Treatment and Utilization. 11(4-5): 33-45.
Wong, P.K., and P.Y. Yuen. 1996. Decolorization and Biodegradation of Methyl Red by Klebsiella Pneumoniae RS-13. Water Research. 30(7): 1736-1744.
Yunus, S.M., H.M. Hamim, O.M. Anas, S.N. Aripin, and S.M. Arif. 2009. Mo (VI) Reduction to Molybdenum Blue by Serratia Marcescens Strain Dr. Y9. Polish Journal of Microbiology. 58(2): 141-147.
Zeng, G.-M., L. Tang, G.-L. Shen, G.-H. Huang, and C.-G. Niu. 2004. Determination of Trace Chromium (VI) by an Inhibition-Based Enzyme Biosensor Incorporating an Electropolymerized Aniline Membrane and Ferrocene as Electron Transfer Mediator. International Journal of Environmental Analytical Chemistry. 84(10): 761-774.
Zhai, X.-W., Y.-L. Zhang, Q. Qi, Y. Bai, X.-L. Chen, L.-J. Jin, X.-G. Ma, R.-Z. Shu, Z.-J. Yang, and F.-J. Liu. 2013. Effects of Molybdenum on Sperm Quality and Testis Oxidative Stress. Systems Biology in Reproductive Medicine. 59(5): 251-255.
Zhang, Y.-L., F.-J. Liu, X.-L. Chen, Z.-Q. Zhang, R.-Z. Shu, X.-L. Yu, X.-W. Zhai, et al. 2013. Dual Effects of Molybdenum on Mouse Oocyte Quality and Ovarian Oxidative Stress. Systems Biology in Reproductive Medicine. 59(6): 312-318.
Mohd Shukri Shukor1, Khan Aftab3, Masdo Norazlina2, Halmi Mohd Izuan Effendi4*, Abdullah Siti Rozaimah Sheikh4 and Shukor Mohd Yunus3
1 Snoc International Sdn Bhd, Lot 343, Inland Port, 71800, Negeri Sembilan, Malaysia
2 Biotechnology Research Centre, MARDI, P. O. Box 12301, 50774 Kuala Lumpur, Malaysia
3 Department of Biochemistry, Faculty of Biotechnology and Biomolecular Sciences, Universiti Putra Malaysia, UPM 43400 Serdang, Selangor, Malaysia
4 Department of Chemical and Process Engineering, Faculty of Engineering and Built Environment, Universiti Kebangsaan Malaysia, 43600 UKM Bangi, Selangor, Malaysia
*Corresponding author. E-mail: zuanfendi88@gmail.com, zuanfendi@ukm.edu.my
Total Article Views

