
Indigenous Saccharomyces cerevisiae Strains from Coconut Inflorescence Sap: Characterization and Use in Coconut Wine Fermentation
Nichanun Udomsaksakul, Kentaro Kodama, Somboon Tanasupawat, and Ancharida Savarajara*Published Date : 2018-07-01
DOI : https://doi.org/10.12982/CMUJNS.2018.0016
Journal Issues : Number 3 , July-September 2018
ABSTRACT
This study isolated Saccharomyces cerevisiae strains from coconut inflorescence sap and evaluated their suitability for coconut wine fermentation. Twenty-six S. cerevisiae strains were isolated from coconut inflorescence sap in Thailand. They were identified based on their morphological, physiological, and biochemical characteristics, as well as by sequence analysis of the D1/D2 region of the large-subunit ribosomal RNA gene. All 26 isolates grew at 37˚C and 11 strains (42%) produced ethanol at more than 50 g/L from 180 g/L glucose containing medium at 30˚C. Sensory evaluation using a nine-point hedonic scale of the coconut wines fermented by five selected yeast strains from 23˚Brix coconut juice showed no significant differences (p = > 0.05) in the color and clarity except for the wine fermented by strain NL010, whose flavor profile and overall acceptance were rated exceptionally high. Thus, coconut inflorescence sap is a potential source of autochthonous S. cerevisiae strains for coconut wine fermentation.
Keywords: Saccharomyces cerevisiae, Coconut inflorescence sap, Wine fermenting, Ethanol
INTRODUCTION
Coconut inflorescence sap is the exudate obtained from the unopened inflorescence of coconut palm (Cocos nucifera). It is a yellowish brown, clear liquid of pH 7.0 that contains 12-15% (w/w) sucrose; approximately 0.23% (w/w) protein; 0.02% (w/w) fat; trace amounts of glucose, fructose, maltose, and raffinose; and is rich in Na and K ions (Kalaiyarasi et al., 2013). Coconut inflorescence sap is rapidly fermented by a heterogeneous population of microorganisms that originate from the environment of the coconut inflorescence. More than 50% of the microorganisms isolated from fermenting coconut inflorescence sap can produce ≥ 9% (v/v) ethanol in batch cultivation using coconut inflorescence sap containing 15–18% (w/v) sucrose (Wijesinghe and Samarajeewa, 1988). Wellala et al. (2004) isolated 12 yeasts from naturally fermenting coconut inflorescence sap in Sri Lanka and reported that all isolates were identified as members of the Saccharomyces genus (species not determined). These Saccharomyces isolates could be categorized into three groups based on the shape and size of the single cells.
Saccharomyces cerevisiae has long been used to produce alcoholic beverages and as baker’s yeast (Bekatorou et al., 2006). Today, S. cerevisiae strains are also used to produce single cell proteins (SCP), ethanol, acetic acid, biocontrol agents, and recombinant vaccines, among others (Ghosh, 2011). The use of indigenous wine yeasts selected from each wine production region is widespread (Lopes et al., 2007). These indigenous yeasts are presumed to be more competitive than commercial yeasts, because they are better adapted to the ecological and technological conditions of their own region. Therefore, they have the potential to dominate fermentation and become the most important biological agent in making wine (Senses-Ergul and Ozbas, 2016). The use of autochthonous yeast strains can contribute to promoting or retaining the natural S. cerevisiae biodiversity (Capece et al., 2016).
The objective of this research was to explore coconut inflorescence sap for indigenous S. cerevisiae strains that have the desired characteristics for coconut wine fermentation.
MATERIALS AND METHODS
Coconut inflorescence sap and yeast isolation
Fifty-six samples of coconut inflorescence sap, freshly harvested from coconut trees, were collected from Samut-Songkram province, Thailand and kept at 4 ºC for no longer than 12 h before use. One mL of the coconut inflorescence sap was inoculated into 5 mL of medium containing 2% (w/v) glucose, 0.3% (w/v) peptone, 0.3% (w/v) yeast extract, 1% (w/v) chloramphenicol, and 3% (v/v) ethanol at pH 5.6 per 16 × 150 mm test tube and incubated in an anaerobic pouch (Kenki, Mitubishi Gas Chemicals., Inc. Tokyo, Japan) at 30 ºC for 5 d. To screen for strong fermenting S. cerevisiae, the obtained culture was purified by the streak plate method on 10% glucose agar medium (10% (w/v) glucose, 0.3% (w/v) peptone, 0.3% (w/v) yeast extract, 1% (w/v) chloramphenicol, 2% (w/v) agar, pH 5.6) and incubated under anaerobic conditions, as above. The resultant single colony was transferred onto YM agar (1% (w/v) glucose, 0.5% (w/v) peptone, 0.3% (w/v) yeast extract, 0.3% (w/v) malt extract, 2% (w/v) agar, pH 5.6) and incubated at 30 ºC under aerobic conditions for 2 d. The obtained culture was kept at 4 ºC for further studies.
Identification of yeast isolates
Conventional identification. Isolated yeasts were identified based on their morphological, physiological, and biochemical characteristics according to the methods described in Kurtzman et al. (2011). Cultures grown on YM agar at 30 ºC for seven days were examined for ascospore production. Formation of pseudohyphae was determined by the Dalmau plate culture method at 25 ºC for 7-14 days using cornmeal agar. Vegetative cell, ascospore, and colony morphologies of YM agar grown cultures were observed and compared to those of the Saccharomyces cerevisiae type strain. Carbon assimilation was determined using the API kit ID 32 C (Biomerieux, France). Two-day-old cells suspended in ultra-pure water at a final MacFarland concentration of 2 (250 μL) were inoculated into C-medium, transferred (135 μL) into well (included in the API kit), and incubated at 30 ºC. The cell turbidity was monitored at 24, 48, and 72 h after incubation.
Carbon fermentation was performed using basal yeast fermentation broth (0.45% (w/v) yeast extract, 0.75% (w/v) peptone, 1% (w/v) bromothymol blue) containing 2% (w/v) of the specific carbon source (glucose, galactose, sucrose, maltose, lactose, raffinose, or trehalose). The ability to ferment the carbon source was examined anaerobically from gas formation in a Durham tube and color change of bromothymol blue (Wickerham, 1951).
Molecular identification. One loopful of yeast cells suspended in 200 μL lysis buffer (100 mM Tris-Cl (pH 8.0), 30 mM EDTA (pH 8.0), 0.5% (w/v) SDS) was boiled for 15 min in a boiling water bath, mixed with 200 μL of 2.5 M potassium acetate (pH 7.5), incubated for 1 h in ice-water, and then centrifuged (18,000xg, 4 ºC, 5 min). The supernatant was extracted twice with 100 μL of chloroform; isoamyl alcohol (24:1) and DNA was precipitated by ethanol precipitation method. The DNA precipitate was dried at 37 ºC, dissolved in sterile water (30 μL). The resultant DNA solution (10-20 μg /μL) was used as a DNA template for PCR amplification using the F63 (5’-GCATATCAATAAGCGGAGGAAAAG-3’) and LR3 (5’-GGTCCGTGTTTCAAGACG-3’) primers and thermal cycling at 94 ºC for 3 min followed by 35 cycles of 94 ºC for 20 s, 52 ºC for 30 s, and 72 ºC for 30 s, and then a final 72 ºC for 5 min. The PCR product (600 bp) was purified using a Gel/PCR DNA fragment extraction kit (PCR clean up: Genaid Biotech Co. Ltd., Taipei, Taiwan), and then direct DNA sequenced commercially at Macrogen Inc., Korea. Each obtained DNA sequence was compared pairwise by BLASTn search and aligned with sequences retrieved from GenBank using the CLUSTALX ver 1.8 multiple alignment program (Thompson et al., 1997). A phylogenetic tree of representative strains of the S. cerevisiae isolates was constructed by Kimura’s model using the neighbor-joining (NJ) method (Saitou and Nei, 1987) with bootstrap random resampling from 1,000 replicates. Sequences determined in this study were deposited at DDBJ (DNA Data Bank of Japan), with their accession numbers shown in Figure 1.
Determination of ethanol production by S. cerevisiae isolates
A single colony was inoculated into 50 mL of fermentation medium (18% (w/v) glucose, 0.45% (w/v) yeast extract, 0.75% (w/v) peptone, pH 5.6) in a 250-mL Erlenmeyer flask and incubated at 30 ºC and 200 rpm for 24 h. The obtained culture was then transferred at 1% (v/v) into 50 mL of the same medium, incubated under the same condition for 24 h, and used as the inoculum. The inoculum was inoculated at 10% (v/v) into the fermentation medium (42.5 mL) in a 50-mL Erlenmeyer flask and incubated at 30 ºC under an oxygen-limited condition for 48 h. The oxygen-limited condition was performed by cultivating 47.5 mL of the inoculated medium in a 50-mL Erlenmeyer flask without shaking; the cotton plug of the culture flask was tightly wrapped with parafilm. After 48 h the culture was centrifuged at 4 ºC and 16700xg for 5 min; the supernatant was analyzed for ethanol by gas chromatography (Hewlett-Packard, HP5890 series, USA) using a Porapak QS (cabowax 20 M) column (2 m × 0.32 m) at an oven temperature of 175 °C and a flame ionization detector at 150 °C. Helium at a flow rate of 35 mL/min was used as the carrier gas (Jutakanoke et al., 2012). The S. cerevisiae TISTR 5596 strain was used as a reference ethanol producing (control) strain. Those S. cerevisiae strains that produced a high ethanol level relative to the control strain were selected for studying coconut wine production.
Coconut wine fermentation
High ethanol producing S. cerevisiae strains were grown on a YM agar slant at 30 ºC for 30 h, eluted with 5 mL of coconut juice (8.0 ºBrix, pH 5.6), and inoculated into 45 mL of coconut juice in a 250-mL Erlenmeyer flask and incubated at 30 ºC and 200 rpm for 24 h. One mL of this culture was transferred into 50 mL of coconut fermentation medium (20% (w/v) coconut sugar in coconut juice, 23 ºBrix, pH 5.6) and incubated at 30 ºC under a static condition for 8 d; the S. cerevisiae cells were then removed by centrifugation (4 ºC, 16,120x g, 5 min). The supernatant was filtered through 0.45 μm membrane, determined for total soluble solid (TSS) level (ºBrix) by hand refractometer, total acidity (as tartaric acid) by the AOAC (2000) method, and ethanol concentration by ebulliometer (Per Vinum J. Salleron Dujardin, Paris). Coconut wine that contained more than 9% (v/v) alcohol was selected for sensory testing.
Sensory test
Coconut wine with an alcohol content of more than 9% (v/v) was presented to a trained panel of 15 randomly selected 20-25-year-old men and women who regularly drank wine. The panelists were asked to evaluate the coconut wine for color, clarity, odor, flavor, and overall acceptance using a nine-point hedonic scale, where 9 = extremely like, 8 = very much like, 7 = moderately like, 6 = slightly like, 5 = neither like nor dislike, 4 = slightly dislike, 3 = moderately dislike, 2 = very much dislike, and 1 = extremely dislike. The data were statistically analyzed by ANOVA and the significance of means using Duncan’s multiple range test (DMRT) accepting significance at the P <0.05 level.
RESULTS
Yeast isolation and identification
Among the yeasts isolated, 26 isolates showed circular colonies with a smooth margin and cream color of 1-3 mm diameter. Cells were spherical to ovoid in shape and reproduced by multilateral budding. The vegetative cell transformed into asci containing 1-4 spherical and smooth ascospores that did not liberate. All of them grew at 37 ºC, but only one isolate, NL009, could grow at 40 ºC. Isolate NL010 produced rudimentary pseudohyphae after incubation at 25 ºC for 7 d (Figure 2). Analysis of their 26S rRNA (D1/D2) sequence revealed a 99.8-100% match to the nucleotide of S. cerevisiae NRRL Y-12632NT (AY048154) (Table 1). Their morphological, physiological, and biochemical characteristics (Table 2) were consistent with S. cerevisiae; the NJ phylogenetic tree also placed all strains within this species (Figure 1).
Table 1. Isolate number, nearest relative, 26S rRNA gene (D1/D2 domain) sequence identity and GenBank/DDBJ accession number of the S. cerevisiae strains.
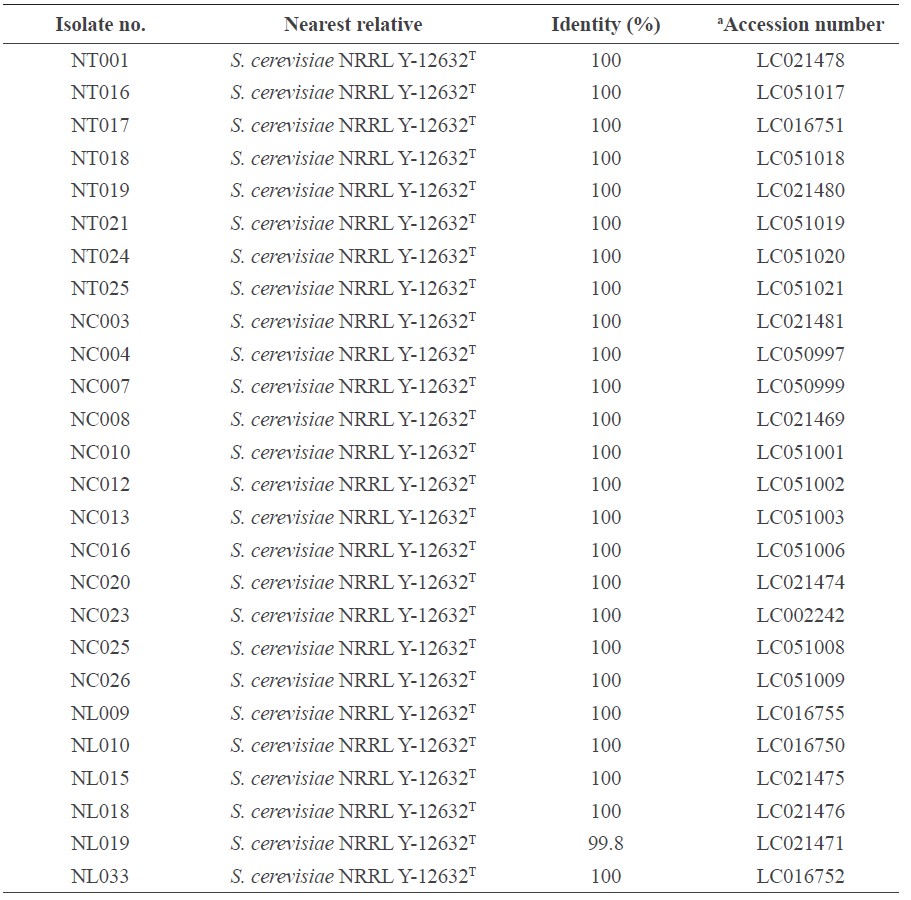
Note: aAccession number: LSU rRNA (D1/D2) sequences determined in this study and deposited at the DDBJ (DNA Data Bank of Japan) gene databank in Japan.
Table 2. Morphological, physiological, and biochemical characteristics of the four S. cerevisiae groups.

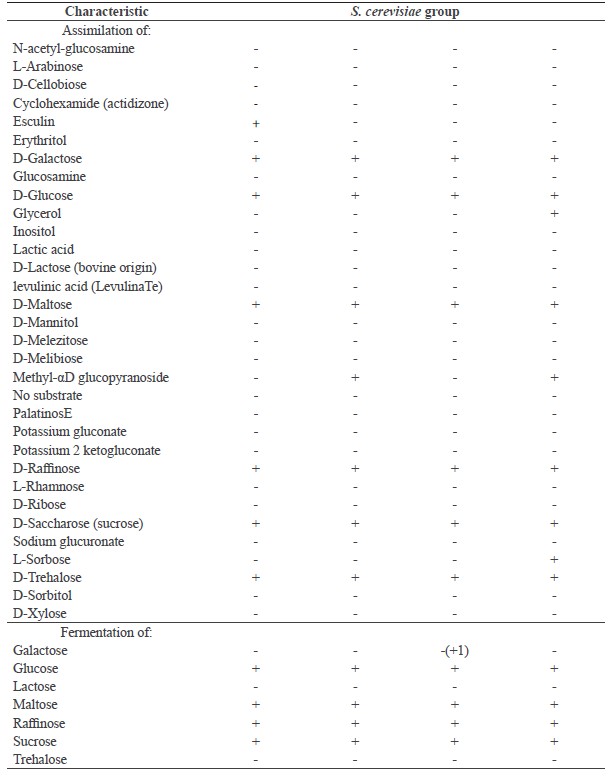
Note: A, strains NL009, NT001, NT016, NT019, NC010, NC012, NC023, NC025 and NC026; B, strains NL033, NT017, NC013 and NC020; C, strains NL015, NL018, NL019, NC003, NC004, NC007, NC008, NT018, NT021, NT024, NT025 and NC016; D, strain NL010. SS, spherical and smooth. Numbers in parentheses indicate the number of strains showing the reaction. Data represent the results from 3 independent assays.
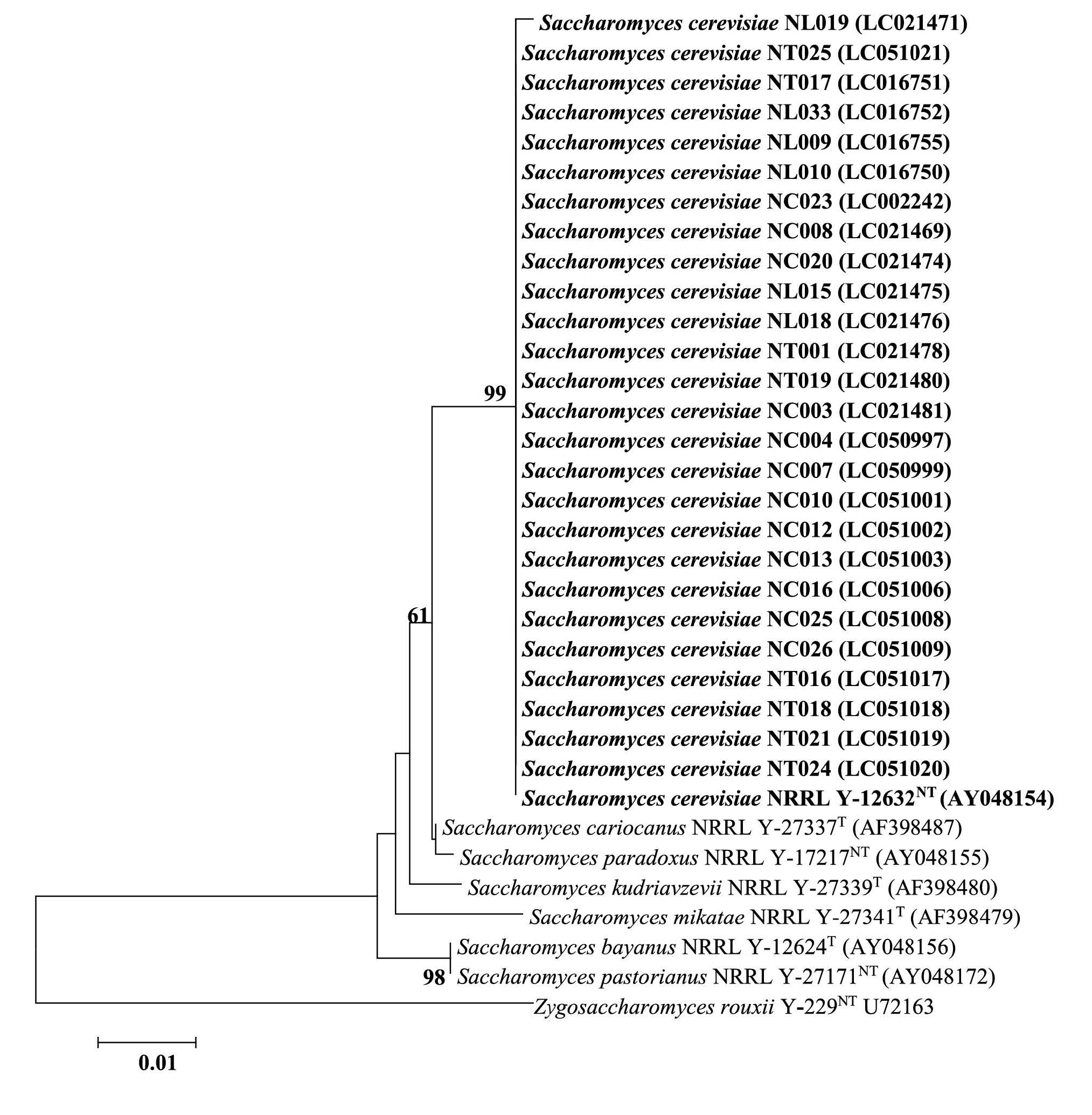
Figure 1. Phylogenetic tree constructed by the NJ method based on 600 bp of the 26S rRNA gene (D1/D2 domain) sequences.
Note: Bold type indicates the position of the reference S. cerevisiae strain (NRRLY-12632NT) and those strains of this study. GenBank/DDBJ accession codes are given in parenthesis after
the species/strain designation. Numbers at nodes represent the percentages from 1,000 replicate bootstrap resamplings when over 50%. Scale bar represents the number of substitutions.
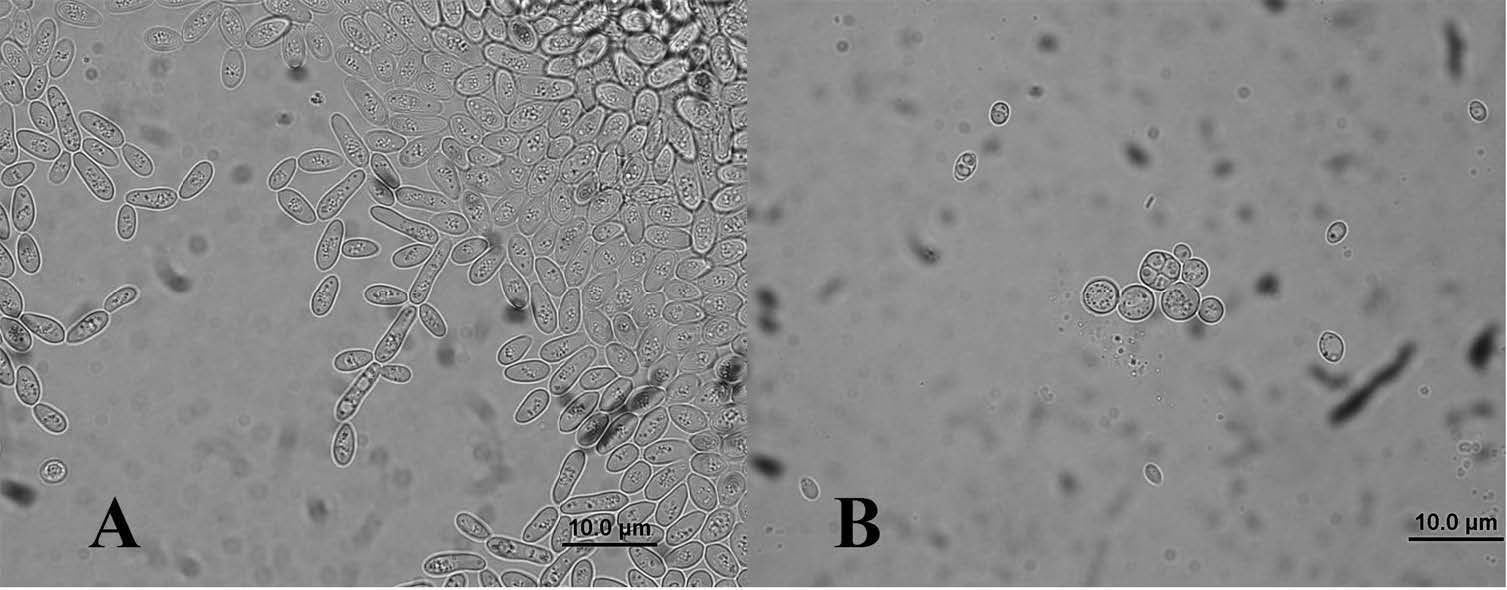
Figure 2. (A) Pseudohyphae cells and (B) ascospore of S. cerevisiae strain NL010 (100X magnification). Images shown are representative of those seen from at least 6-7 such fields of view.
Based on these results, they were identified as S. cerevisiae (Pretorius et al., 1999). The 26 strains of S. cerevisiae were divided into four groups (A–D) based upon their morphology, carbon assimilation, and fermentation differences (Table 2).
Group A contained nine strains (NL009, NT001, NT016, NT019, NC010, NC012, NC023, NC025, and NC026) that all assimilated glucose, galactose, sucrose, raffinose, maltose, trehalose, and esculin, and fermented glucose, sucrose, maltose, and raffinose. Only strain NL009 could grow at 40 ºC (Table 2).
Group B contained four strains (NL033, NT017, NC013, and NC020) that were different from Group A in their ability to assimilate methyl-αD glucopyranoside, but not esculin (Table 2).
Group C contained 12 strains (NL015, NL018, NL019, NC003, NC004, NC007, NC008, NC016, NT018, NT021, NT024, and NT025) that differed from Groups A and B in their ability to assimilate esculin and methyl-αD glucopyranoside, respectively. However, one strain (NL019) could also ferment galactose (Table 2).
Group D contained one strain (NL010) that differed from Groups A-C in the presence of rudimentary pseudohyphae and its ability to assimilate glycerol and L-sorbose, and from Groups A and C by its ability to assimilate methyl-αD glucopyranoside (Table 2).
Ethanol production
From the four groups of S. cerevisiae, 11 representative strains – NL009, NL010, NL015, NL018, NL019, NL033, NT001, NT017, NT019, NC008, and NC023 – were selected and examined for their ethanol production in 18% (w/v) glucose containing medium at 30 ºC for 48 h. All of the representative strains selected produced ethanol at more than 50 g/L with an ethanol productivity of more than 1.04 g/L/h (Figure 3). Strain NC008 produced the highest maximum ethanol yield at 67.22 g/L, which was higher than that produced by the reference S. cerevisiae TISTR 5596 strain (66.52 g/L).
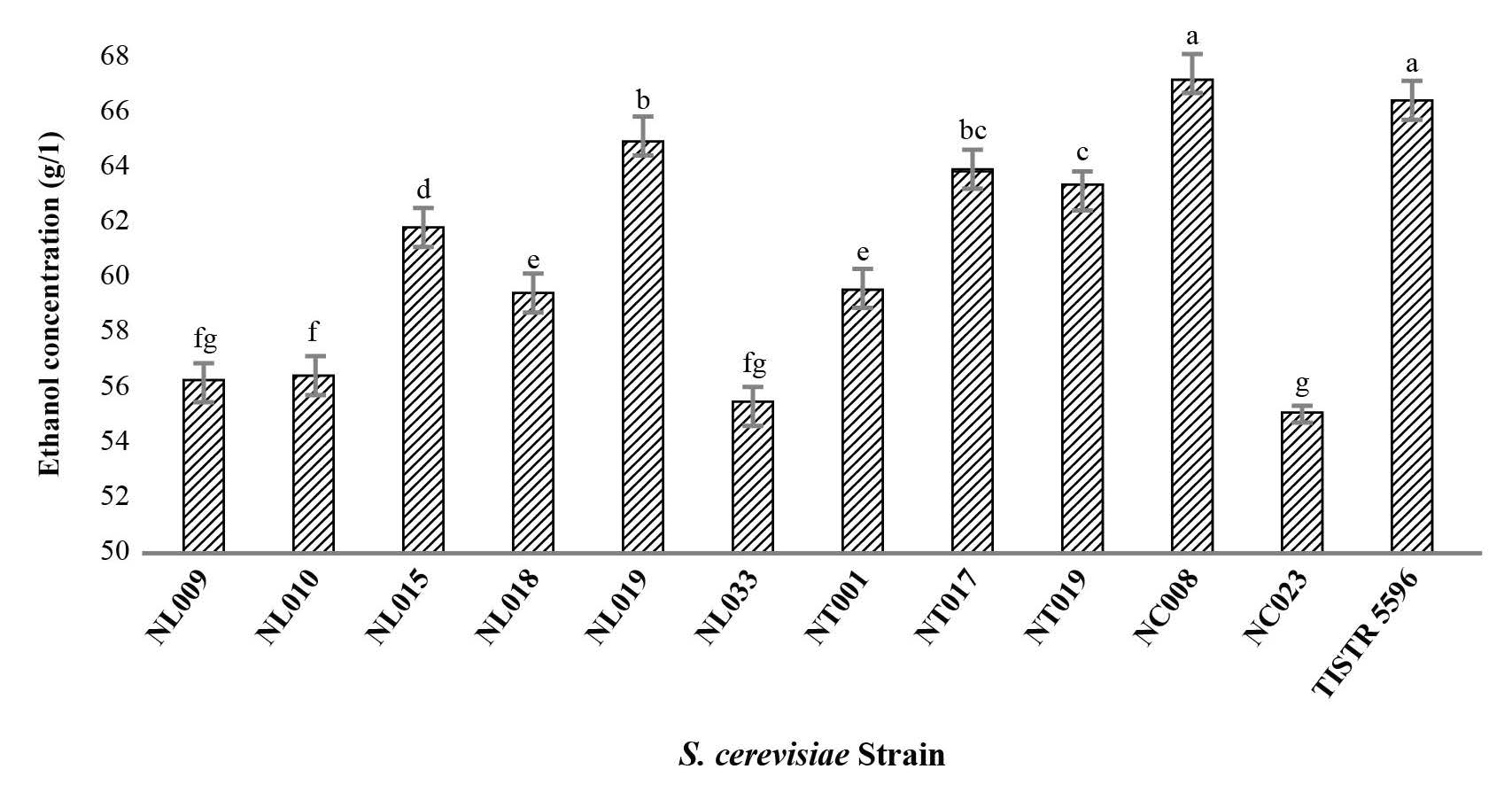
Figure 3. Ethanol production by S. cerevisiae strains at 30 ºC.
Coconut wine fermentation and sensory test
After eight days of fermentation, the coconut wine fermented by strains NL009, NL010, NL033, NC023, and NT017 all contained more than 9% (v/v) alcohol with a pH in the range of 2.9-3.8. Strain NL010 gave the highest alcohol content wine at 10.3% (v/v), with the other three strains being slightly lower at 9.3-9.4% (v/v), and the lowest TSS content except for that produced by strain NL009 (Table 3).
Table 3. The total soluble solid, pH, total acidity, and ethanol concentration of coconut wine fermented by the selected five S. cerevisiae strains.
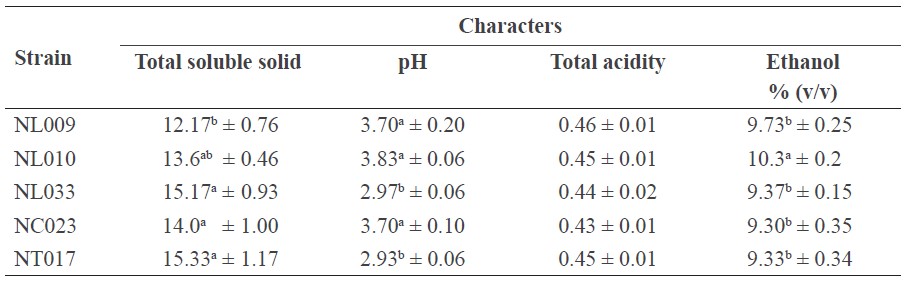
Note: Data are shown as the mean ± SD, derived from three independent replicates. Means within the same column followed by a different letter are significantly different (P < 0.05; DMRT).
Fifteen panelists evaluated these coconut wines fermented by the five selected S. cerevisiae strains (NL009, NL010, NL033, NC023, and NT017) for their sensory qualities. The result indicated that the wines did not markedly differ, although the wine fermented by strain NL010 had the highest overall acceptance and best flavor, while that fermented by strain NL009 had the best odor (Table 4).
Table 4. Sensory test of coconut wine fermented by the five selected S. cerevisiae strains.
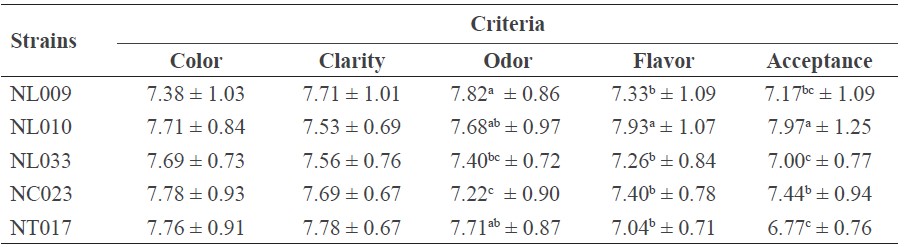
Note: Data are shown as the mean ± SD, derived from three independent replicates. Means within the same column followed by a different letter are significantly different (P < 0.05; DMRT).
DISCUSSION
From the 26 strains of S. cerevisiae isolated from coconut inflorescence sap, only one strain (NL009) could grow at 40 ºC. This result agreed well with Cassey and Ingledew (1996) in that yeast cell viability decreased with increasing temperature, and D’Amore et al. (1989) that growth and budding of yeast were fast at 30-37 ºC, but growth decreased at higher temperatures. Fermenting yeast tolerant to high temperature has several advantages – it minimizes contamination risk, decreases the cost of cooling during fermentation, and reduces the energy required for proper agitation rate due to the decrease in fermentation broth viscosity at high temperature.
Carbon assimilation is an important criterion in the identification of yeasts that utilize organic carbon compounds as an energy source for growth. All the isolated S. cerevisiae strains were able to ferment glucose, sucrose, maltose, and raffinose; could not ferment lactose and trehalose; and had a variable response to galactose. This result agreed with Dash et al. (2015) and Jimoh et al. (2011, 2012) that most yeasts that fermented glucose, galactose, maltose, sucrose, and raffinose were S. cerevisiae.
The maximum alcohol concentration of the coconut wine fermented by the five strains of S. cerevisiae selected was 10.3% (v/v). This accorded with Nzabuheraheza and Nyiramugwera (2014) who fermented must containing 18º Brix sugar that yielded a wine with 10% (v/v). The wine fermented by strain NL010 had the highest overall acceptance and best flavor according to the trained sensory panel.
Studies have shown that changes in the composition and sensory quality of coconut wine can indicate the variety of the S. cerevisiae strains. In particular, distinct strains of S. cerevisiae induced considerably different flavor characteristics when the same must is fermented (Mauriello et al., 2009). Currently, commercially available strains of S. cerevisiae are used to produce the majority of wine. However, many researchers and wine producers opt for the autochthonous strains of S. cerevisiae as the starter culture (Capece et al., 2014) because indigenous S. cerevisae strains adapt better to the micro-area conditions in certain wine producing regions. Moreover, locally chosen yeast strains and strain-specific metabolic characteristics can be used together, beneficially affecting the quality of the wine produced (Capece et al., 2012).
CONCLUSION
An indigenous coconut inflorescence sap S. cerevisiae, strain NL010, was isolated that could produce a coconut wine of 10.3% (v/v) alcohol with a satisfactory overall acceptance and flavor profile, as determined by a sensory panel based on a nine-point hedonic scale.
REFERENCES
AOAC (Association of Official Analytical Chemistry) 2000. Official Methods of Analysis. 17th Ed. Arlington, Virginia, Gaithersburg, M.D. USA.
Bekatorou V., Psarianos C., and Koutinas, A.A. 2006. Production of food grade yeasts. Biotechnology. 44: 407-415.
Cassey, G., and Ingledew, W.M. 1996. Ethanol tolerance in yeasts. Critical Review in Microbiology. 13: 219-280. https://dx.doi.org/ 10.3109/10408418609108739
Capece, A., Romaniello, R., Siesto, G., and Romano, P. 2012. Diversity of Saccharomyces cerevisiae yeasts associated to spontaneously fermenting grapes from an Italian "heroic vine-growing area". Food Microbiology. 31(2): 159-166. https://dx.doi.org/10.1016/j.fm.2012.03.010. Epub 2012 Mar 29.
Capece, A., Romaniello, R., Pietrafesa, R., and Romano, P. 2014. Indigenous Saccharomyces cerevisiae yeasts as a source of biodiversity for the selection of starters for specific fermentations. BIO web of Conferences. 3: 02003. https://dx.doi.org/10.1051/bioconf/20140302003.
Capece, A., Granchi, L., Guerrini, S., Mangani, S., Romaniello, R., Vincenzini, M., and Romano, P. 2016. Diversity of Saccharomyces cerevisae strains isolated from two Italian Wine-Producing Regions. Frontier in Microbiology. 3: 1018. https://dx.doi.org/10.3389/fmicb.2016.010118.
D’Amore, T., Russell, I., and Stewart, G.G. 1989. Sugar utilization by yeast during fermentation. Journal of Industrial Microbiology. 4: 315-324. https://dx.doi.org/10.1007/BF01577355.
Dash P.K., Mishra J., Patnaik S.C., Swain M.R., and Thatoi, H.N. 2015. Characterization, identification and comparative evaluation of bioethanol tolerance and production capacity of isolated yeasts strains from fermented date palm sap (Toddy). Malaysian Journal of Microbiology. 11: 223-230. http://dx.doi.org/10.21161/mjm.57213.
Ghosh, S. K. 2011. Study of yeast flora from fruit of Syzygium cumini (Linn) skeel. Agriculture and Biology Journal of North America. 2: 1166-1170. https://doi.org/10.5251/abjna.2011.2.8.1166.1170
Grimwood, B.E., and Ashman, F. 1975. Coconut palm production: Their process in developing countries. Food and Agriculture Organization. 189-190.
Jimoh, S.O., Ado, S.A., Ameh, J.B., and Whong, C.M.Z. 2011. Characteristics and diversity of yeast in locally fermented beverages sold in Nigeria. Research Journal of Biological Sciences. 6(8): 389-392. https://doi.org/10.3923/rjbsci.2011.389.392
Jimoh, S.O., Ado, S.A., Ameh, J.B., and Whong, C.M.Z. 2012. Characteristics and diversity of yeast in locally fermented beverages sold in Nigeria. World Journal of Engineering and Pure and Applied Science 2(2): 40.
Jutakanoke, R., Leepipatpiboon, N., Tolieng, V., Kitpreechavanich, V., Srinorakutara, T., and Akaracharanya, A. 2012. Sugarcane leaves: Pretreatment and ethanol fermentation by Saccharomyces cerevisiae. Biomass and Bioenergy. 39: 283-289. http://dx.doi. org/10.1016/j.biombioe.2012.01.018.
Kalaiyarasi K., Sangeetha, K., and Rajarajan S. 2013. A comparative study on the microbial flora of the fresh sap from cut inflorescence and fermented sap (toddy) of Borrassus flabellifer Linn. (palmyrah tree) and of Cocos nucifera Linn. (coconut tree) to identify the microbial fermenters. International Journal of Research in Pure and Applied Microbiology. 3: 43-47.
Kurtzman, C.P., Fell J.W., and Boekhout T. 2011. The Yeasts: A Taxonomic Study, 5th Ed. Elsevier Science Publisher, USA.
Lopes, C.A., Rodriguez, M.E., Sangorrin, M., Querol, A., and Caballero, A.C. 2007. Patagonian wines: the selection of an indigenous yeast starter. Journal of Industrial Microbiology & Biotechnology. 34: 539-546. https://doi.org/10.1007/s10295-007-0227-3
Mauriello, G., Capece, A., D’Auria, M., Garde-Cerdan, T., and Romano, P. 2009. SPME-GC method as a tool to differentiate VOC profiles in Saccharomyces cerevisiae wine yeasts. Food Microbiology. 26(3): 246-252. https://dx.doi.org/10.1016/j.fm.2009.01.003.Epub 2009 Jan 20.
Nzabuheraheza, F.D., and Nyiramugwera, A.N. 2014. Golden wine produced from mixed juices of passion fruit (Passiflora edulis), mango (Mangifera indica) and pineapple (Ananas comosus). African Journal of Food Agriculture, Nutrition and Development. 14: 9104-9116.
Pretorius, I.S., Van der Westhuizen, T.J., and Augustyn, O.P.H. 1999. Yeast biodiversity in vineyards and wineries and its importance to the South African industry- a review. South African Journal of Enology and Viticulture. 20: 61-64. http://dx.doi.org/10.21548/20-2-2234
Saitou, N., and Nei, M. 1987. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 4: 406-425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Senses-Ergul, S., and Ozbas, Y.Z. 2016. Characterization of some indigenous Saccharomyces cerevisiae isolates obtained during vinification of ‘Kalecik karasi’ and ‘emir’ grapes grown in central Anatolia. Ciencia e Tecnica Vitivinicola. 31(2)51-62. http://dx.doi.org/10.1051/ ctv/20163102051.
Thompson, J.D., Gibson T.J., Plewniak, F., Jeanmougin, F., and Hoggins, J.D. 1997. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acid Research. 24: 4876-4882. http://dx.doi.org/ 10.1093/nar/25.24.4876.
Wellala, C.K.D., Gunawardhane, M.H.W., Wijeratne, M.C.P., and Illeperuma, C.K. 2004. Fermentation of rice using yeast isolated from coconut and palmyrah sap. Agricultural Research. 16: 114-120.
Wickerham, L.J. 1951. Taxonomy of the Yeasts. Technical Bulletin of the U.S. 1029: 1-55.
Wijesinghe, D.G.N.G., and Samarajeewa, U. 1988. Screening yeasts from coconut inflorescence sap for continuous alcoholic fermentation. Food Microbiology. 5:119-123. https://doi.org/10.1016/0740-0020(88)90009-3
Nichanun Udomsaksakul1, Kentaro Kodama2, Somboon Tanasupawat3, and Ancharida Savarajara4*
1 Program in Biotechnology, Faculty of Science, Chulalongkorn University, Bangkok 10330, Thailand
2 The Institute of Biotechnology and Genetic Engineering, Chulalongkorn University, Bangkok 10330, Thailand
3 Department of Biochemistry and Microbiology, Faculty of Pharmaceutical Sciences, Chulalongkorn University, Bangkok 10330, Thailand
4 Department of Microbiology, Faculty of Science, Chulalongkorn University, Bangkok 10330, Thailand
*Corresponding author. E-mail: sanchari@chula.ac.th
Total Article Views

