
Quantification of Ochratoxin A–Producing Fungi in Coffee Products Using Quantitative PCR
Jakapan Potipun, Wijittra Ruaylarb, Rungrote Nilthong and Amorn Owatworakit*Published Date : 2018-01-01
DOI : https://doi.org/10.12982/CMUJNS.2018.0004
Journal Issues : Number 1 , January-March 2018
ABSTRACT
Ochratoxin A (OTA) is a polyketide mycotoxin that is produced by Aspergillus and Penicillium. Food contaminated with OTA poses health risks and is a food-safety challenge. Quantitative polymerase chain reaction (qPCR) has been used to identify non-toxigenic and toxigenic strains from coffee samples using polyketide synthase (pks), the OTA synthesis gene. In this research, Aspergillus carbonarius (ochratoxin-producing strain) and A. flavus (non-ochratoxin-producing strain) were used to amplify a 141 bp fragment of the pks gene. The 141 bp PCR product was successfully cloned into TOPO®TA plasmid. Subsequently, ten-fold dilutions of plasmid DNA were used to generate the standard curve by plotting the threshold cycle against log DNA concentration using qPCR. Further, fungal DNA contamination was quantified in 11 samples of roasted coffee using qPCR. All 11 coffee samples were accepted as safe, since the fungal genomic DNA contamination was less than 3.85 x 103 copies. Therefore, this research suggested that qPCR is a fast and accurate method to detect and quantify OTA-producing fungi in coffee products. Thus, we successfully developed a system to quantify fungal contamination in coffee.
Keywords: Aspergillus carbonarius, Ochratoxin A, Coffee, Quantitative PCR, Polyketide synthase (pks) gene
INTRODUCTION
Ochratoxin A (OTA) is a pentaketide mycotoxin that exhibits immunosuppressive, teratogenic, and carcinogenic properties. OTA is also a potent nephrotoxin and the possible causative agent of Balkan endemic nephropathy in humans (Leong et al., 2006). The toxin can be found in a broad range of processed and unprocessed foodstuffs, including coffee. Coffee is one of the most popular drinks in the world and a valuable primary product. The presence of OTA in the various stages of coffee processing is of great concern. The major source of OTA in coffee is fungi of the genus Aspergillus (Pardo et al., 2004; Velmourougane et al., 2011). Specifically, Aspergillus carbonarius is one of the main species responsible for production and accumulation of the toxin in coffee. This fungus has also been reported to have the highest ochratoxigenic potential (Mulè et al., 2006).
Given the negative effects of OTA, it is important to have a reliable method for detecting and quantifying OTA-producing fungi in foodstuffs. Conventional techniques are not accurate. Moreover, identifying Aspergillus species based on morphological characters is difficult and requires taxonomic expertise. Furthermore, spore isolation and enumeration may introduce bias against slow-growing species (Selma et al., 2008). Previous studies have shown that detecting OTA-producing Aspergillus using PCR methods is specific, sensitive, rapid, and easy to automate. (Atoui et al., 2007).
Specifically, quantitative polymerase chain reaction (qPCR) is a powerful tool that combines fluorescent dyes and sequence-specific primers to monitor accumulation of PCR product during the procedure. Furthermore, it does not require other post-amplification procedures, such as gel electrophoresis. Thus, this technique is highly reliable, sensitive, and suitable for high throughput analysis (Mulè et al., 2006). Quantitative PCR has been used to quantify OTA-producing fungi in many agricultural products, including wine, cereal grain, tea, and coffee (Atoui et al., 2007). Common targets include polyketide synthase (pks), a gene involved in synthesis of secondary metabolites and OTA, as well as internal transcribed spacer (ITS) rRNA (Sartori et al., 2006; Atoui et al., 2007).
This work aimed to develop a method for quantifying A. carbonarious, an OTAproducing fungus, in coffee products using qPCR to provide a useful food-safety tool.
MATERIALS AND METHODS
Coffee samples and fungal strains
Eleven roasted coffee products were purchased from coffee companies in Chiang Rai province. A. carbonarius TISTR3214 (OTA-producing strain) and A. flavus TISTR3130 (non-OTA–producing strain) were obtained from the Thailand Institute of Scientific and Technological Research (TISTR) culture collection and used as reference strains. Fungal strains were grown at 30°C on potato dextrose agar for 7 days. Subsequently, their spores were collected in 0.1% (v/v) between 80 and cultivated in 100 ml of potato dextrose broth at 30°C without shaking for 3 days.
Fungal genomic DNA extraction from reference strains
One hundred mg of fungal mycelia from A. carbonarius were frozen in liquid nitrogen before nucleic acid extraction. Genomic DNA was extracted using the DNA secure Plant Kit (TIANGEN) following manufacturer’s instructions. The quality and quantity of DNA were estimated by using the OD260/280 ratio and 3% agarose gel electrophoresis.
Fungal DNA extraction from coffee samples
Five g of roasted coffee beans were soaked in 10 mL lysis buffer (66mM EDTA, 33mM Tris, 3.3% Triton X-100, 1.65M Guanidinium-HCl, 0.825M NaCl, 6% Polyvinyl pyrrolidone-40T, pH=7.9), added to 10 mL ddH2O, and then shaken vigorously by hand for 1 min. One mL of supernatant was taken and mixed with 0.4 mL of absolute ethanol. DNA was extracted using the Tiangen® DNA extraction kit. Subsequently, DNA solutions were kept at - 20°C.
PCR reaction
The total volume of the PCR reaction was 20 μl reaction, which contained: 2 μl of Taq polymerase buffer 10x, 2 μl of dNTP, 1μl of each primer, 1 μl of Taq, about 30 ng of genomic DNA, topped up to 20 μl with dH2O. Reaction conditions were: 94°C for 4 min; followed by 28 cycles of 94°C for 40s, 65°C for 40s, and 72°C for 40s; and final extension at 72°C for 10 min. The primer pair, OTAF (5’-AATATATCGACTATCTGGACGAG CG-3’) and OTAR (5’-GAAGCCCTCTGCGATCTCCC-3’) was used to amplify a 141 bp fragment of the pks gene. The amplified products were examined by 3% agarose gel electrophoresis (Atoui et al., 2007).
Cloning of pks gene and analysis
The TOPO TA Cloning® Kit (Invitrogen) was used for cloning. The PCR products were inserted into the vector and the bacterial E. coli DH5α cells were transformed using the heat shock method. Subsequently, the recombinant cells were cultured on Luria-Bertani medium for 24 hr. Positive colonies were picked and plasmid isolation was performed using TIANprep Rapid Mini Plasmid Kit® (TIANGEN).
DNA sequencing analysis
The extracted plasmids were bidirectionally sequenced using OTA primers (First BASE Laboratories, Malaysia).
Real-time qPCR reaction
Quantitative PCR reactions were run in triplicate. Each reaction well contained 10 μl of template DNA, 5 μl of SYBRR® Green I, and 0.5 μL of each forward and reverse OTA primer. Real-time qPCR was conducted using the following cycling conditions: 50°C for 2 min, 95°C for 10 min, followed by 40 cycles of 95°C for 15s, and 60°C for 60s. The DNA standard curve was generated by plotting the threshold cycle (Ct) versus the logarithm of known DNA concentrations using a series of 10-fold dilutions of a plasmid containing the pks gene (ranging from 3x108-3x103 DNA copies). Quantification of A. carbonarius pks gene was performed by running the DNA from the coffee samples in parallel with the serial dilution standard.
RESULTS
Amplification of pks gene from fungal strains
A fragment of the pks gene was amplified only from A. carbonarious using specific OTA primers. The PCR product was of the expected size of 141 bp (Figure 1). A. flavus, the non OTA-producing fungus gave a negative result. These results showed that the primers used for PCR amplification were specific for targeting pks gene in A. carbonarious.
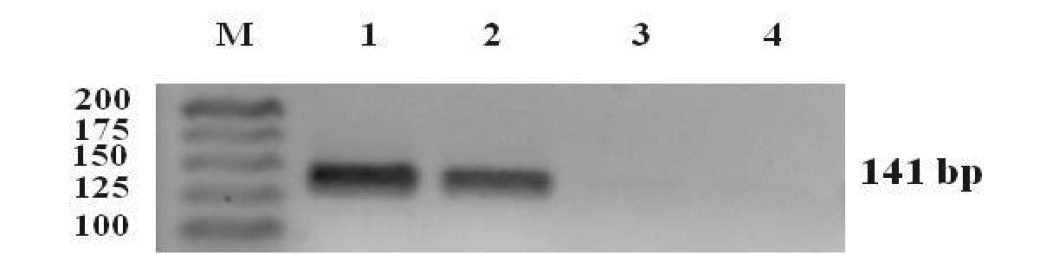
Figure 1. 3% agrose gel electrophoresis of PCR products from the fungal strains amplified with OTA primers. Lane M contains the 25 bp DNA ladder; Lanes 1 and 2 contain the pks gene of A. carbonarious; Lanes 3 and 4 are A. flavus.
OTA cloning and sequencing analysis
A 141 bp fragment of the pks gene from extracted plasmids was amplified using PCR (Figure 2.). The obtained pks gene sequences were then compared with the database in the National Center for Biotechnology and Information (NCBI) using the BLAST search tool. The fragment was confirmed as pks based on 98% identity with the already published A. carbonarius pks sequence (Atoui et al., 2006).
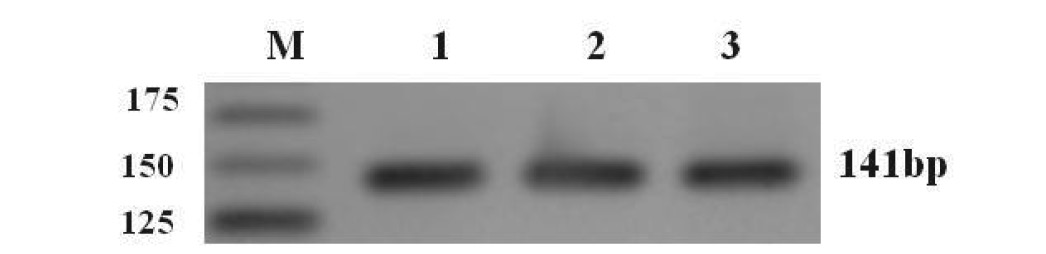
Figure 2. 3% agrose gel electrophoresis of the 141 bp pks fragment amplified with OTA primers. Lane M contains the 25 bp DNA ladder; Lanes 1, 2, and 3 contain the TOPO plasmid with the 141 bp fragment of pks gene.
Amplification of OTA gene from coffee samples
All 11 coffee samples were positive for the pks gene as indicated by the presence of the specific PCR product amplified with OTA primers (Figure 3). This result demonstrated fungal contamination in commercially available coffee in Chiang Rai.
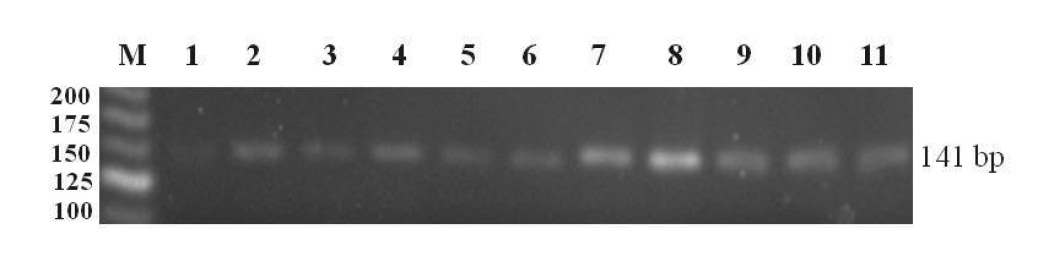
Figure 3. 3% agrose gel electrophoresis of the 141 bp fragment of pks gene amplified with OTA primers. Lane M contains the 25 bp DNA ladder; Lanes 1-11 show the PCR products obtained from coffee samples.
Quantification of A. carbonarious using qPCR
For every qPCR run, a standard curve was constructed using triplicate reactions of 10-fold dilutions of A. carbonarius plasmid DNA ranging from 3x103-3x108 DNA copies conjugated with SYBR® Green I dye. The standard curve was obtained by plotting the threshold cycle (Ct) corresponding to the logarithm of the plasmid DNA concentration of each dilution (Table 1). The linear correlation coefficient of the standard curve was R2=0.99, and melt curve analysis indicated the specificity and accuracy of the PCR-based quantification (Figure 4).
The Ct values of the 11 coffee samples obtained from the real-time PCR system ranged from 24 to 31 (Table 2), indicating that only a small amount of fungal DNA was present in the coffee samples used in this experiment. Only coffee sample no. 8 could be quantified containing 3.85x103 DNA copies. The rest of the samples could not be reliably estimated using the equation from the standard curve (Figure 4B). This indicated that the DNA copy number of OTA-producing fungi must be less than 3.85x103. Consistently, the total number of fungal species in roasted coffee using culture-based approaches was 10 and 5x103 CFU/5g (data not shown).
Table 1. Ct values corresponding to the standard curve obtained using SYBR-Green I with genomic DNA from A. carbonarius.
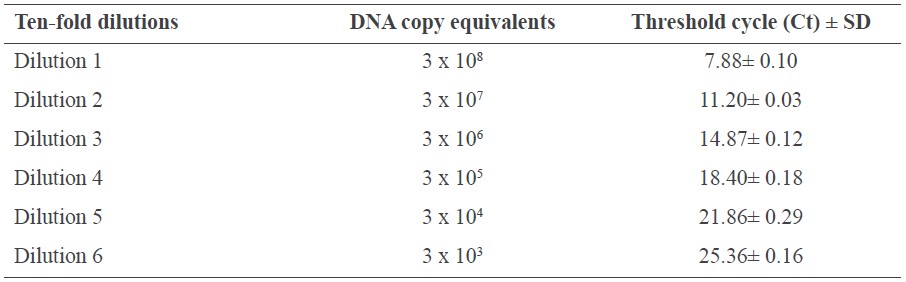
Figure 4. (A) A plot of relative fluorescence units corresponding to cycle number of the amplification. (B) Standard curve showing the log10 DNA amount (ng) vs. threshold cycle of the real-time PCR method for the 10-fold serial dilutions of A. carbonarius.
Table 2. Threshold cycle (Ct) value of 11 coffee samples obtained from real-time PCR.
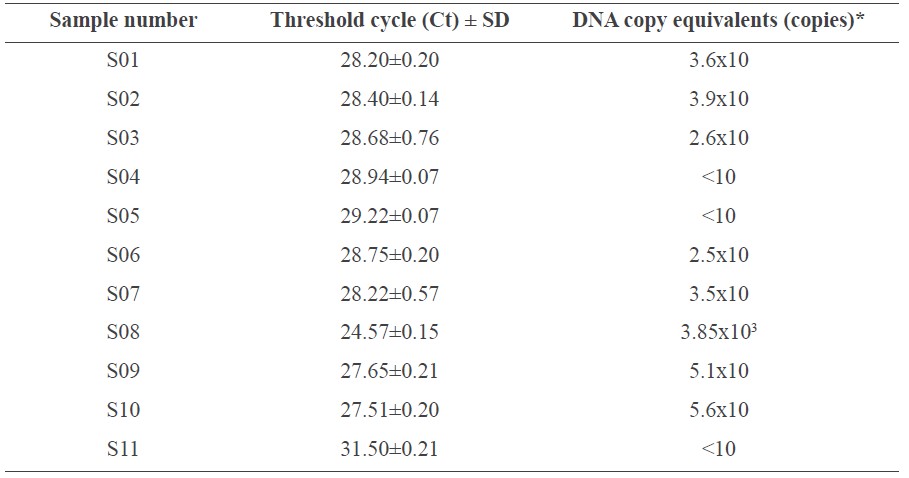
Note: The values were derived from averaging three replicates. *The pks gene copy number equivalents of A. carbonarius from coffee samples were calculated using a standard equation.
DISCUSSION
Aspergillus species produce ochratoxin A, which is a toxic secondary metabolite. This mycotoxin is nephrotoxic and carcinogenic and has been detected in cereal and other food commodities, such as coffee (Bucheli et al., 1998). The PCR reaction has been used to detect and quantify mycotoxin-producing Aspergillus species. In this study, we used specific OTA primers to detect pks, the OTA synthesis gene, in fungal strains (Figure 1). Previous research has shown that amplification of pks gene in Aspergillus species using these primers was specific to A. carbonarius only, as indicated by the specific 141 bp PCR product (Atoui et al., 2007).
Specificity of the primer pair OTAF/OTAR was comfirmed using PCR assay. The resulting amplicon was then cloned into a plasmid. The sequence of the amplicon had a 98% identity to the published A. carbonarius polyketide synthase 5 (accession number: HM026487) in GenBank (Atoui et al., 2006).
In order to examine the presence of OTA-producing fungi in the 11 DNA samples extracted from coffee, PCR reactions were performed using OTAF/OTAR primers to amplify a 141 bp fragment of the pks gene. The pks gene was detected in all coffee samples (Figure 3 and 4).
This study shows that qPCR is a useful tool for detecting and quantifying A. carbonarius in coffee samples or other food products. It is necessary to use rapid and specific methods for early detection of OTA-producing fungi, especially when foods are involved (Dao et al., 2005). Recently, a rapid, specific, and sensitive qPCR assay for detecting and quantifying A. carbonarius on coffee was developed. In our experiment, we generated the standard curve, which was obtained by plotting the Ct value versus the log of the concentration of each DNA dilution (Figure 4) with the regression coefficient (R2) greater than 0.99. The qPCR method was highly sensitive; the smallest amount of fungal DNA that it detected was 3.85x103 DNA copies. To test the ability of the qPCR method to quantify OTA-producing fungi in coffee samples, the Ct values of the samples (Table. 1) were compared with the standard curve. Given the detection limits of real-time PCR systems, very low concentrations of DNA can lead to difficulty in detecting fluorescence due to the low amount of SYBR intercalating with the sample. The maximum Ct value for detection in our work was 25. Our data are in agreement with previous results that also used qPCR assays for A. carbonarius quantification in artificially inoculated samples (Atoui et al., 2007). The A. carbonarius DNA concentration was quantified in coffee samples and was between 10-14-10-9g DNA/5g of coffee sample.
CONCLUSION
This research successfully identified the pks gene of A. carbonarius in Arabica coffee by using specific OTAF/OTAR primers. We developed a real-time PCR to quantify fungal DNA concentration in Arabica coffee. In addition, this technique was specific, as well as, sensitive. The described method could be a useful tool for screening, monitoring, and detecting contamination with OTA-producing fungi in commercial food products. Further study is required to quantify the concentration of OTA contamination in coffee products.
ACKNOWLEDGEMENTS
This research was supported by a grant from Mae Fah Luang University and presented at the 28th Annual Meeting of the Thai Society for Biotechnology and International Conference. We also thank Dr. Khanobporn Tangtrakulwanich and Dr. Gentekaki Eleni for help with the manuscript.
REFERENCES
Atoui, A., Dao, H.P., Mathieu, F., and Lebrihi, A. 2006. Amplification and diversity analysis of ketosynthase domains of putative polyketide synthase genes in Aspergillus ochraceus and Aspergillus carbonarius producers of ochratoxin A. Molecular Nutrition and Food Research. 50(6): 488-493. https://doi.org/10.1002/mnfr.200500165
Atoui, A., Mathieu, F., and Lebrihi, A. 2007. Targeting a polyketide synthase gene for Aspergillus carbonarius quantification and ochratoxin A assessment in grapes using real-time PCR. International Journal of Food Microbiology. 115(3): 313-318. https://doi.org/10.1016/j.ijfoodmicro.2006.11.006
Bucheli, P., Meyer, I., Pittet, A., Vuataz, G., and Viani, R. 1998. Industrial storage of green robusta coffee under tropical conditions and Its impact on raw material quality and ochratoxin A content. Journal of Agricultural and Food Chemistry. 46(11): 4507-4511. https://doi.org/10.1021/jf980468+
Dao, H.P., Mathieu, F., and Lebrihi, A. 2005. Two primer pairs to detect OTA producers by PCR method. International Journal of Food Microbiology. 104(1): 61-67. https://doi.org/10.1016/j.itfoodmicro.2005.02.004
Leong, S.L., Hocking, A.D., Pitt, J.I., Kazi, B.A., Emmett, R.W., and Scott, E.S. 2006. Australian research on ochratoxigenic fungi and ochratoxin A. International Journal of Food Microbiology. 111, Supplement 1(0): S10-S17. https://doi.org/10.1016/j.itfoodmicro.2006.02.005
Mulè, G., Susca, A., Logrieco, A., Stea, G., and Visconti, A. 2006. Development of a quantitative real-time PCR assay for the detection of Aspergillus carbonarius in grapes. International Journal of Food Microbiology. 111(SUP.1): S28-S34. https://doi.org/10.1016/j.ijfoodmicro.2006.03.010
Pardo, E., Marı́n, S., Solsona, A., Sanchis, V., and Ramos, A.J. 2004. Modeling of germination and growth of ochratoxigenic isolates of Aspergillus ochraceus as affected by water activity and temperature on a barley-based medium. Food Microbiology. 21(3): 267-274. https://doi.org/10.1016/j.fm.2003.09.001
Selma, M.V., Martínez-Culebras, P.V., and Aznar, R. 2008. Real-time PCR based procedures for detection and quantification of Aspergillus carbonarius in wine grapes. International Journal of Food Microbiology. 122(1-2): 126-134. https://doi.org/10.1016/j.ijfoodmicro.2007.11.049
Sartori, D., Furlaneto, M.C., Martins, M.K., Ferreira de Paula, M.R., Pizzirani-Kleiner, A.A., Taniwaki, M.H., and Fungaro, M.H.P. 2006. PCR method for the detection of potential ochratoxin-producing Aspergillus species in coffee beans. Research in Microbiology. 157(4): 350-354. https://doi.org/10.1016/j.resmic.2005.09.008
Velmourougane, K., Bhat, R., Gopinandhan, T.N., and Panneerselvam, P. 2011. Management of Aspergillus ochraceus and Ochratoxin-A contamination in coffee during on-farm processing through commercial yeast inoculation. Biological Control. 57(3): 215-221. https://doi.org/10.1016/j.biocontrol.2011.03.003
Jakapan Potipun, Wijittra Ruaylarb, Rungrote Nilthong and Amorn Owatworakit*
School of Science, Mae Fah Luang University, Chiang Rai 57100, Thailand
*Corresponding author. E-mail: amorn@mfu.ac.th
Total Article Views

