
Exploring the Relationship Between Oral Pathogens and Gingival Health in Adults with Gingivitis and Mild Periodontitis: An Observational Cross-sectional Study
Woo Hyun Shim, Jusun Song, Ki Hwan Kim, and Sanghun Lee*Published Date : November 22, 2024
DOI : https://doi.org/10.12982/NLSC.2025.010
Journal Issues : Number 1, January-March 2025
Abstract Gingivitis, characterized by inflammation of the gingiva, is prevalent globally and can progress to more severe periodontal diseases if untreated. Dysbiosis, an imbalance in the oral microbiota, plays a critical role in the progression of gingivitis. This study investigates the relationship between ten pathogenic bacteria and clinical indices (Plaque Index, Gingival Index, Bleeding on Probing, and Probing Pocket Depth) used in the diagnosis and assessment of gingivitis in adults under the hypothesis that specific oral pathogens correlate with these indices.
A cross-sectional study was conducted on 130 adults diagnosed with gingivitis or mild periodontitis at Myongji Hospital, South Korea. Supragingival plaque samples were collected and analyzed using PowerCheck™ Periodontitis Pathogens Multiplex Real-time PCR to identify the prevalence of ten periodontitis-related pathogens. Statistical analyses, including multiple linear regression, were performed to determine associations between bacterial prevalence and clinical indices, adjusting for age and sex. Significant associations were found between several periodontal pathogens and clinical indices.
Fusobacterium nucleatum exhibited a strong correlation with Plaque Index (P-value=5.61×10-5) and Gingival Index (P-value=4.02×10-4), suggesting its relevance in the inflammatory profile of gingivitis and mild periodontitis. Other pathogens also demonstrated varying degrees of association with the clinical indices.
The study highlights the value of integrating bacterial analysis with conventional diagnostic approaches for comprehensive assessment and management of gingivitis. The findings suggest that Fusobacterium nucleatum may contribute to early inflammatory processes in gingival health, potentially informing targeted therapeutic strategies in periodontal care.
Keywords: Gingivitis, Periodontal pathogens, Fusobacterium nucleatum, Plaque index, Gingival index
Funding: This research was supported by grants of the Myongji Hospital (grant number: 2203-09-02).
Citation: Shim, W. H., Song, J., Kim, K. W., and Lee, S. 2025. Exploring the relationship between oral pathogens and gingival health in adults with gingivitis and mild periodontitis: An observational cross-sectional study. Natural and Life Sciences Communications. 24(1): e2025010.
INTRODUCTION
Gingivitis, a prevalent and mild form of periodontal disease, is characterized by inflammation of the gingiva, the part of the gum surrounding the base of the teeth (Kim and Amar 2006). The global prevalence of gingivitis is estimated to be 45-50%, with higher rates observed among high-income and elderly population (Nazir et al. 2020). Gingivitis, while initially limited to gum inflammation, may progress to periodontitis if left untreated. Periodontitis, in turn, can lead to tooth loss and other serious oral health outcomes (Elter et al. 2004; Kim and Amar 2006). Periodontal disease has been increasingly recognized for its associations with various systemic diseases, highlighting the complex interplay between oral health and overall systemic well-being. Research indicates that conditions such as diabetes mellitus, cardiovascular disease, rheumatoid arthritis, and inflammatory bowel diseases share common inflammatory pathways with periodontal disease, suggesting a bidirectional relationship (Nabila et al. 2023; Martinez-Garcia and Hernandez-Lemus 2024). Therefore, early detection and effective treatment of periodontal diseases are crucial not only for maintaining periodontal health but also for preventing these systemic conditions (Isola et al. 2023).
According to the 2018 periodontal disease classification (Chapple et al. 2018), gingivitis is diagnosed when gingival bleeding exceeds 10% of the sites examined, with bleeding as the primary sign of inflammation. Also, gingivitis is closely linked to the presence and activity of specific bacterial communities in the oral cavity (Paster et al. 2001; Aas et al. 2005; Darveau 2010). The balance between beneficial and harmful bacteria in the mouth is crucial for maintaining oral health (Darveau 2010). Poor oral hygiene can disrupt this balance, leading to a condition known as dysbiosis, where pathogenic bacteria dominate (Darveau 2010). This microbial imbalance is a key factor in the transition from gingivitis to more severe periodontal diseases, such as periodontitis (Na et al. 2020; Ali and Taha 2024).
Periodontal disease progression is largely driven by specific bacterial communities within the oral microbiome, where shifts toward dysbiosis create a pathogenic environment (Socransky et al. 1998; Bartold and Van Dyke 2019). Among the diverse oral bacteria, we selected ten key pathogens — Aggregatibacter actinomycetemcomitans, Porphyromonas gingivalis, Tannerella forsythia, Treponema denticola, Prevotella intermedia, Fusobacterium nucleatum, Parvimonas micra, Filifactor alocis, Porphyromonas endodontalis, and Treponema socranskii — due to their demonstrated roles in periodontitis progression. Direct associations between these bacteria and early signs of gingivitis still remain underexplored. By analyzing these specific pathogens in individuals with gingivitis and mild periodontitis, our study aims to bridge this knowledge gap and enhance the clinical understanding of microbiota's role in early-stage periodontal disease management. Here, we conducted a detailed comparison of these bacteria with traditional diagnostic methods such as the Plaque Index (PI), Gingival Index (GI), Bleeding on Probing (BOP) index, and Probing depth of periodontal pockets (Rahardjo et al. 2005; Checchi et al. 2009). These methods are routinely employed by dental professionals to assess the presence and severity of gingivitis, relying primarily on visual and physical indicators of the disease. However, identifying specific pathogenic bacteria provides deeper insights into the microbial origins of gingivitis, offering potential for more precise and targeted treatment strategies (Bartold and Van Dyke 2019). Integrating bacterial analysis with established clinical indices is crucial for enhancing diagnostic accuracy and improving therapeutic outcomes in the management of gingivitis. This approach bridges the gap between microbiological findings and conventional diagnostic practices, aiming to advance periodontal disease management and patient care.
MATERIAL AND METHODS
Study participants
In the study, we hypothesized that specific oral pathogens correlate with gingivitis severity and the primary outcome was the association between bacterial prevalence and gingival indices. The inclusion criteria required adult male and female participants aged 19 to under 65 who expressed concern regarding their gingival health, had a diagnosis of gingivitis with potential progression to periodontitis, or exhibited mild periodontitis, excluding cases with chronic periodontitis. Eligible participants were required to have a BOP score exceeding 10% and at least one tooth with a periodontal pocket probing depth (PPD) between 3 and 5 mm. Participants also needed to display clinical symptoms of gingivitis or mild periodontitis and possess a minimum of 20 remaining teeth.
Exclusion criteria applied to individuals who had undergone scaling within the previous month, received periodontal treatment within the past six months, presented serious pathological conditions in oral soft tissues, or had five or more teeth necessitating immediate intervention for dental caries. Additionally, individuals were excluded if they presented with significant cardiovascular, immune, infectious, or neoplastic diseases, or mental health conditions such as schizophrenia, depression, substance abuse, or alcohol addiction. Participants were also ineligible if they had taken drugs influencing periodontal health (e.g., phenytoin, calcium channel blockers, cyclosporine, NSAIDs, or coumadin) or antibiotics or adjunctive periodontal therapy within the preceding month. Further exclusion applied to those consuming alcohol above daily limits (over 40g for men and 20g for women), current smokers, and those who had ingested health supplements or foods specifically targeting gum health or related products within the past month.
Ethical considerations
This observational cross-sectional study was conducted in accordance with ethical guidelines and approved by the Institutional Review Board at Myongji Hospital (Approval No. 2022.10-014). All participants in the study provided informed consent prior to participation after receiving sufficient explanation. This study involved a total of 130 participants and was conducted at Myongji Hospital from March 17, 2023, to December 11, 2023. We adhered to the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) guidelines for observational cross-sectional studies, with all relevant aspects reported accordingly (supplementary materials).
Clinical index measurements for gum health
Clinical indices were measured at the lingual and mesial-distal sides of four teeth (Rt. Top #11, Lt. Top #26, Rt. Bot #43, and Lt. Bot #34) by two dentists. The teeth were chosen for accessibility and balance across quadrants according to the previous studies that identified 16 types of periodontal bacteria in the four sites of gingival sulcus of two maxillary teeth and two mandibular teeth (Kang et al. 2020). In cases of missing teeth, the following teeth were examined: Lt. Top #21, Rt. Top #16, Lt. Bot #33, Rt. Bot #44. The measurements were conducted using a 0.4-mm periodontal probe (WHO-style probe, Hu-Friedy Inc, Chicago, IL, USA), with calibration performed using a standardized calibration block. Regular calibration checks were conducted throughout the study to ensure consistent accuracy.
1) PI (Silness and Loe 1964): The plaque index was evaluated on the buccal surface of the maxillary and mandibular first molars, as well as the buccal surface of the maxillary central incisors and the lingual surface of the mandibular central incisors. Plaque was assessed using an explorer and scores were assigned as follows:
0: No plaque present
1: Thin film of plaque detectable by running the explorer across the tooth surface
2: Moderate accumulation of plaque along the gingival margin with no plaque visible between teeth
3: Abundant plaque covering the gingival margin with plaque visible between teeth
2) GI (Loe and Silness 1963): The evaluation method for the GI involved dividing each tooth into four areas: buccal, mesial, distal, and lingual. Gingival inflammation was observed and scored as follows:
0: Normal gingiva
1: Mild inflammation, slight color change, slight swelling, no bleeding tendency upon probing
2: Moderate inflammation, redness, swelling, color change of the gingiva, bleeding upon probing
3: Severe inflammation, significant redness and swelling, presence of ulceration, continuous bleeding
3) Gingival Recession (GR) (Smith 1997): The distance (mm) from the midbuccal cementoenamel junction (CEJ) to the gingival margin was measured using a periodontal probe for all teeth. In cases with prosthetic restorations, the distance from the restoration margin to the gingival margin was measured.
4) PPD (Anderson et al. 1991) and Clinical Attachment Level (CAL): PPD (mm) was recorded using a periodontal probe graduated in 1mm increments, measuring the distance from the base of the gingival sulcus to the gingival margin, while CAL (mm) was measured from the base of the gingival sulcus to the CEJ.
5) BOP (Furuta et al. 2011): BOP was recorded as positive (+) if bleeding occurred within 30 seconds of probing the buccal, mesial, or distal surfaces of each tooth, and negative (-) if no bleeding occurred. BOP was expressed as a percentage of sites showing bleeding upon probing.
Periodontal pathogens
Subgingival plaque samples were collected with a sterile cotton swab from the buccal, lingual, and occlusal surfaces of the teeth used in clinical index measurements (Zheng et al. 2024). Participants abstained from eating for two hours and from brushing their teeth for four hours before sampling collection. Immediately after sampling, the specimens were placed in a collection kit with preservatives (NBgene-GUT kit; Noble Biosciences, Republic of Korea) and stored at -80°C for later analysis. They were sent to the GC Genome Inc. for PCR quantitative measurement for bacterial identification. The bacterial genomic DNA was extracted using MagNA Pure 96 DNA and Viral NA small volume kit (Roche Diagnostics, Germany) according to the manufacturer’s instructions. DNA concentration was determined fluorometrically on the Qubit® 3.0 Fluorometer (Thermo Fisher Scientific, USA) using the QubitTM dsDNA HS Assay Kit. The real-time PCR was carried out using PowerCheckTM Periodontitis Pathogens Multiplex Real-time PCR kit (KogeneBiotech, Seoul, Korea) which was developed for detecting 10 periodontitis-related pathogens including Red complex, Orange complex and Aa-complex (Socransky et al. 1998; Sim et al. 2024). The target genes for each oral bacteria are described in supplementary Table 1. Amplifications were performed with pre-heating and initial melting at 50°C and 95°C for 2 min and 10 min, respectively, followed by 40 cycles of denaturing at 95°C for 15 sec, annealing and extension at 60°C for 1 min. Standard curves for each organism were plotted for each primer-probe set using the Ct (the cycle number at which the threshold fluorescence was reached) values obtained by amplifying successive 10-fold dilutions of a known copies of the bacteria. Real-time PCR was performed and analyzed using the CFX-96 real-time PCR detection system (Bio-Rad, Hercules, CA, USA). To validate specificity, we indirectly assessed it by performing repeated experiments with positive control DNA containing nucleic acids from all target bacterial species with known the quantification cycle (Cq) values. We verified specificity by comparing the observed Cq values for each probe against expected Cq values. Each experimental batch included both positive control DNA and a no-template control. We confirmed that the Cq values for the positive control DNA were within three standard deviations on the internally organized Levey-Jennings Control Chart, while no positive Cq values were observed for the no-template control.
Statistical analysis
Statistical analyses were performed using R (version 4.2), a free package from the R Foundation for Statistical Computing. The sample size of 130 was determined based on the lowest prevalence of bacterial pathogens reported in similar studies, specifically with A. actinomycetemcomitans as a keystone pathogenic bacterium exhibiting a prevalence of approximately 9% among individuals with periodontitis (Scapoli et al. 2015). Using the prevalence, along with a 95% confidence level and a margin of error of ±5%, an initial calculation suggested a minimum of 126 participants. However, the sample size was increased to 130 to account for potential variability in measurements and to enhance the study’s generalizability. Demographic and clinical characteristics of subjects were reported as mean ± standard deviation for continuous variables, and as frequencies and percentages for categorical variables. We applied log10-transformation with raw values of periodontal pathogens determined by PCR. Multiple linear regression was used to determine the association between a clinical index and a periodontal pathogen after adjusting for the covariates, including age and sex. A P-value < 0.05 was considered statistically significant.
RESULTS
Patients’ characteristics
Table 1 presents the demographic and clinical characteristics of the study participants. A total of 130 individuals were included who consisted of 34 men and 96 women (73.85%), with a mean age of 38.754 ± 11.753 years (range: 23─65) and a mean BMI of 22.186 ± 3.077 kg/m2. In comorbidity, only 4 patients had hypertension and 2 ones were under statin therapy for dyslipidemia.
All participants were diagnosed with either gingivitis or mild periodontitis. Clinical measurements for gum health included a mean PI of 1.104 ± 0.316 (range: 1─3), a mean GI of 1.096 ± 0.302 (range: 1─3), a mean GR of 0.048 ± 0.273 (range: 0─3), a mean CAL of 2.812 ± 0.410 (range: 1.75─6.25), a mean BOP of 29.33 ± 13.27 (range: 12.5─75.0), and a mean PPD of 2.764 ± 0.276 (range: 1.75─3.33, Table 1). Among the measurements, age was significantly associated with BOP and PPD (P-value = 2.26×10-2, 8.29×10-4, respectively) while sex showed no significant association with any of the clinical parameters (Table 2).
Periodontal pathogen distributions
Table 1 also details the prevalence of the 10 periodontal pathogens identified in the study. Several pathogens including P. gingivalis, P. intermedia, F. nucleatum, and A. actinomycetemcomitans, exhibited a skewed distribution, even after log10 transformation. Figure 1 illustrates the correlations among these pathogens, revealing notable associations. Specifically, T. forsythia was strongly correlated with T. denticola, F. alocis, and P. endodontalis, while T. denticola was highly correlated with F. alocis and P. endodontalis. Additionally, P. endodontalis showed strong correlations with F. alocis, P. micra, and T. socranskii.
Table 1. Patients’ characteristics, Clinical measurements related to gum health, and periodontal pathogens. Mean ± SD or number (%) is shown.
|
Variables |
Subjects (Number=130) |
|
Clinical characteristics |
|
|
Age (years) |
38.754 ± 11.753 |
|
Female (number) |
96 (73.85) |
|
Body Mass Index (kg/m2) |
22.186 ± 3.077 |
|
Alcohol consumption (number) |
54 (41.53) |
|
Hypertension (number) |
4 (58.46) |
|
Dyslipidaemia (number) |
2 (1.54) |
|
Clinical measurements related to gum health |
|
|
Plaque Index (PI) |
1.104 ± 0.316 |
|
Gingival Index (GI) |
1.096 ± 0.302 |
|
Gingival Recession (GR, mm) |
0.048 ± 0.273 |
|
Clinical Attachment Level (CAL, mm) |
2.812 ± 0.410 |
|
Bleeding on Probing (BOP, %) |
29.33 ± 13.27 |
|
Probing pocket depth (PPD, mm) |
2.764 ± 0.276 |
|
Periodontal pathogens (Log10 DNA copy number) |
|
|
Aggregatibacter actinomycetemcomitans |
0.104± 0.473 |
|
Porphyromonas gingivalis |
0.759 ± 1.466 |
|
Tannerella forsythia |
1.338 ± 1.558 |
|
Treponema denticola |
1.104 ± 1.633 |
|
Prevotella intermedia |
0.857 ± 1.643 |
|
Fusobacterium nucleatum |
0.159 ± 0.743 |
|
Parvimonas micra |
1.440 ± 1.264 |
|
Filifactor alocis |
1.116 ± 1.498 |
|
Porphyromonas endodontalis |
2.151 ± 1.669 |
|
Treponema socranskii |
3.137 ± 1.484 |
Table 2. Linear regression of age and sex for clinical parameters related to gum health.
|
Clinical parameters related to gum health |
Age |
Sex (reference: female) |
|||||
|
Beta |
SE |
P-value |
Beta |
SE |
P-value |
||
|
Plague Index (PI) |
1.96×10-3 |
2.38×10-3 |
0.412 |
9.60×10-2 |
6.35×10-2 |
0.133 |
|
|
Gingival Index (GI) |
2.22×10-3 |
2.28×10-3 |
0.332 |
9.75×10-2 |
6.07×10-2 |
0.111 |
|
|
Gingival Recession (GR) |
3.21×10-3 |
2.06×10-3 |
0.122 |
-1.99×10-2 |
5.49×10-2 |
0.718 |
|
|
Clinical Attachment Level (CAL) |
-1.06×10-2 |
6.99×10-3 |
0.132 |
-1.84×10-1 |
1.86×10-1 |
0.324 |
|
|
Bleeding On Probing (BOP) |
4.57×10-3 |
1.98×10-3 |
2.26×10-2 |
-2.31×10-2 |
5.48×10-2 |
0.662 |
|
|
Probing Pocket Depth (PPD) |
6.88×10-3 |
2.01×10-3 |
8.29×10-4 |
6.91×10-2 |
5.35×10-2 |
0.199 |
|
Note: Results of multiple linear regression analyses to evaluate the influence of age and sex on clinical indices, with significance set at P< 0.05.
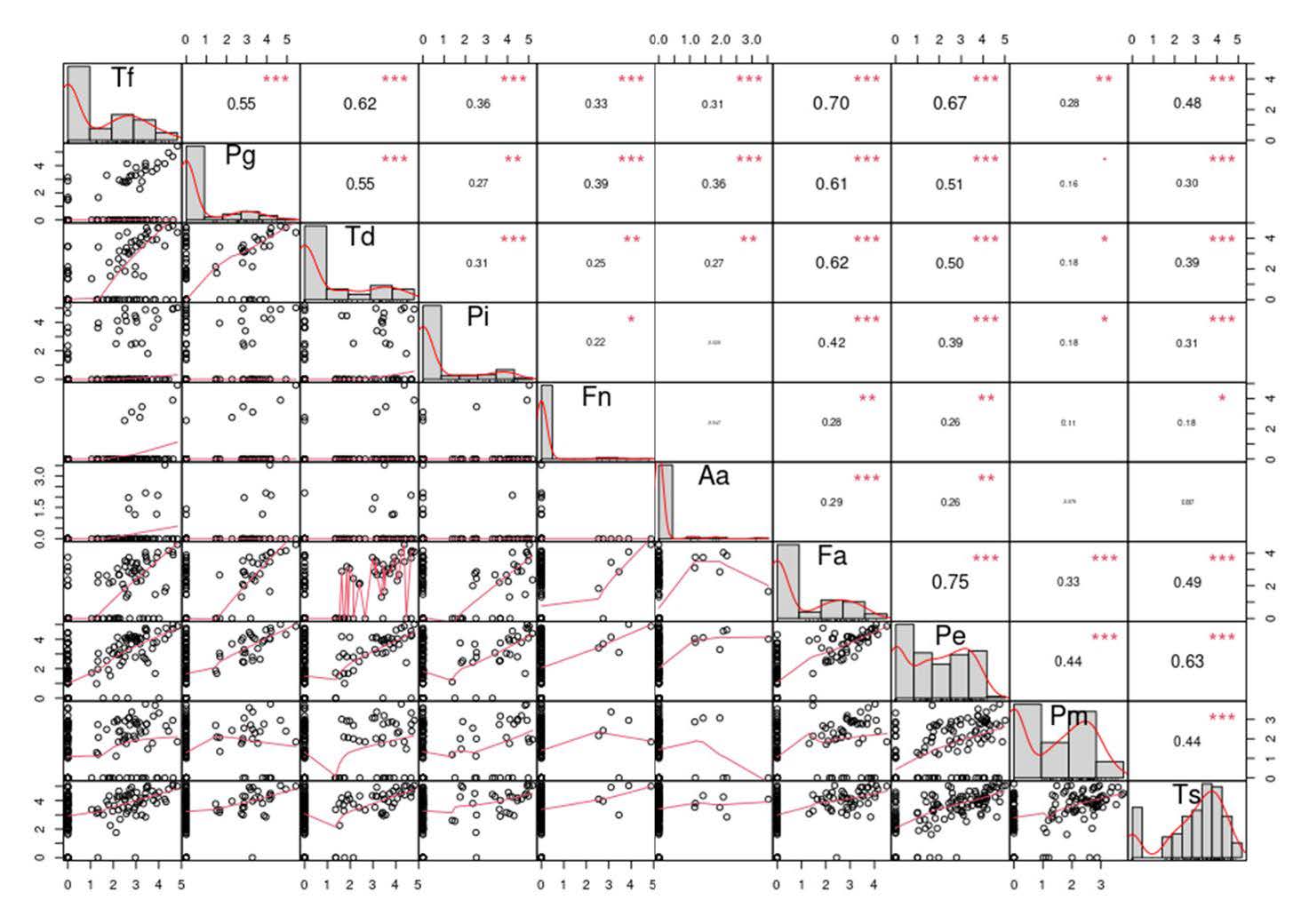
Figure 1. Correlation matrix for all pairs of the observed periodontal pathogens: Aggregatibacter actinomycetemcomitans (Aa), Porphyromonas gingivalis (Pg), Tannerella forsythia (Tf), Treponema denticola (Td), Prevotella intermedia (Pi), Fusobacterium nucleatum (Fn), Parvimonas micra (Pm), Filifactor alocis (Fa), Porphyromonas endodontalis (Pe), and Treponema socranskii (Ts). The lower part shows scatter plots and smoothing splines for each possible pair; the upper part shows the Pearson correlation coefficient (text size proportional to absolute value). The significance of the correlation coefficients at different levels is indicated by the following symbols: 0.05 (*), 0.01 (**) and 0.001 (***).
Periodontal pathogens for clinical index in gum health
Tables 3 to 5 present the associations between clinical gum health measurements and periodontal pathogens, adjusted for age and sex through multiple linear regression. Significant associations were observed between PI, GI, and most of periodontal pathogens, with the exceptions of P. intermedia, A. actinomycetemcomitans, and T. socranskii (Table 3). Notably, F. nucleatum was strongly associated with both PI and GI (P-value=5.61×10-5, 4.02×10-4, respectively). Among all pathogens, only F. nucleatum was significantly associated with GR (P-value=1.28×10-3), whereas no significant association were found between CAL or PPD and any pathogens (Table 4 & 5). Finally, BOP is marginally associated with P. micra and T. socranskii (P-value=0.0610, 0.0451, respectively).
Table 3. Associations of each periodontal pathogen for plaque index (PI) and gingival index (GI).
|
Periodontal pathogens |
Plague Index (PI) |
Gingival Index (GI) |
||||
|
Beta |
SE |
P-value |
Beta |
SE |
P-value |
|
|
Tannerella forsythia |
0.0553 |
0.0176 |
2.03×10-3 |
0.0548 |
0.0167 |
1.37×10-3 |
|
Porphyromonas gingivalis |
0.0663 |
0.0196 |
9.85×10-4 |
0.0635 |
0.0188 |
9.63×10-4 |
|
Treponema denticola |
0.0386 |
0.0169 |
0.0244 |
0.0429 |
0.0161 |
8.69×10-3 |
|
Prevotella intermedia |
0.0207 |
0.0170 |
0.2260 |
0.0180 |
0.0163 |
0.2710 |
|
Fusobacterium nucleatum |
0.1530 |
0.0367 |
5.61×10-5 |
0.1290 |
0.0356 |
4.02×10-4 |
|
Aggregatibacter actinomycetemcomitans |
0.0267 |
0.0589 |
0.6510 |
0.0427 |
0.0562 |
0.4490 |
|
Filifactor alocis |
0.0631 |
0.0182 |
7.05×10-4 |
0.0621 |
0.0173 |
4.85×10-4 |
|
Porphyromonas endodontalis |
0.0439 |
0.0165 |
8.87×10-3 |
0.0434 |
0.0158 |
6.72×10-3 |
|
Parvimonas micra |
0.0538 |
0.0221 |
0.0163 |
0.0579 |
0.0210 |
6.60×10-3 |
|
Treponema socranskii |
0.0298 |
0.0190 |
0.1200 |
0.0271 |
0.0182 |
0.1400 |
Note: Multiple linear regression was used to evaluate the association of PI or GI on specific periodontal pathogens, adjusted for age and sex.
Table 4. Associations of each periodontal pathogen for gingival recession (GR) and clinical attachment level (CAL).
|
Periodontal pathogens |
Gingival Recession (GR) |
Clinical Attachment Level (CAL) |
||||
|
Beta |
SE |
P-value |
Beta |
SE |
P-value |
|
|
Tannerella forsythia |
0.0204 |
0.0156 |
0.1950 |
0.0044 |
0.0534 |
0.9340 |
|
Porphyromonas gingivalis |
0.0086 |
0.0177 |
0.6260 |
0.0308 |
0.0600 |
0.6090 |
|
Treponema denticola |
-0.0075 |
0.0149 |
0.6130 |
0.0208 |
0.0506 |
0.6820 |
|
Prevotella intermedia |
0.0013 |
0.0148 |
0.9280 |
-0.0871 |
0.0495 |
0.0810 |
|
Fusobacterium nucleatum |
0.1070 |
0.0324 |
1.28×10-3 |
-0.1940 |
0.1130 |
0.0887 |
|
Aggregatibacter actinomycetemcomitans |
0.0113 |
0.0509 |
0.8240 |
0.0221 |
0.1730 |
0.8980 |
|
Filifactor alocis |
-0.0130 |
0.0164 |
0.4300 |
-0.0103 |
0.0557 |
0.8540 |
|
Porphyromonas endodontalis |
0.0150 |
0.0146 |
0.3070 |
-0.0352 |
0.0496 |
0.4790 |
|
Parvimonas micra |
0.0195 |
0.0194 |
0.3170 |
-0.0542 |
0.0660 |
0.4130 |
|
Treponema socranskii |
0.0113 |
0.0166 |
0.4980 |
-0.0316 |
0.0563 |
0.5750 |
Note: Multiple linear regression was used to evaluate the association of GR or CAL on specific periodontal pathogens, adjusted for age and sex.
Table 5. Associations of each periodontal pathogen for bleeding on probing (BOP) or probing pocket depth (PPD).
|
Periodontal pathogens |
Bleeding On Probing (BOP) |
Probing Pocket Depth (PPD) |
||||
|
Beta |
SE |
P-value |
Beta |
SE |
P-value |
|
|
Tannerella forsythia |
0.0132 |
0.0151 |
0.3840 |
0.00492 |
0.0154 |
0.749 |
|
Porphyromonas gingivalis |
-0.0045 |
0.0170 |
0.7910 |
0.00077 |
0.0173 |
0.964 |
|
Treponema denticola |
0.0047 |
0.0143 |
0.7430 |
0.01090 |
0.0145 |
0.454 |
|
Prevotella intermedia |
0.0144 |
0.0141 |
0.3110 |
0.00525 |
0.0144 |
0.716 |
|
Fusobacterium nucleatum |
0.0164 |
0.0324 |
0.6130 |
0.02030 |
0.0329 |
0.538 |
|
Aggregatibacter actinomycetemcomitans |
-0.0697 |
0.0485 |
0.1530 |
0.00199 |
0.0496 |
0.968 |
|
Filifactor alocis |
0.0001 |
0.0158 |
0.9950 |
-0.01320 |
0.0160 |
0.410 |
|
Porphyromonas endodontalis |
0.0085 |
0.0141 |
0.5450 |
-0.00384 |
0.0143 |
0.789 |
|
Parvimonas micra |
0.0350 |
0.0185 |
0.0610 |
0.00415 |
0.0190 |
0.828 |
|
Treponema socranskii |
0.0318 |
0.0157 |
0.0451 |
-0.00660 |
0.0162 |
0.684 |
Note: Multiple linear regression was used to evaluate the association of BOP or PPD on specific periodontal pathogens, adjusted for age and sex.
DISCUSSION
Gingivitis is caused by pathogenic bacteria in the oral cavity, with the proliferation and reduction of these microbes influenced by various factors such as nutritional imbalance, stress, oral hygiene, hormones, and smoking (Jenkinson and Lamont 2005; Chattopadhyay et al. 2024). Conventional diagnostic methods for detecting gingivitis and tracking the progression of chronic periodontitis often involve discomfort, inconveniences, and significant time commitment due to frequent dental visits (Calladine et al. 2022). Patients are typically reluctant to seek dental visits unless experiencing discomfort (Armfield et al. 2008). Because clinical pain is not a common symptom in the early stages of gingivitis, most patients only seek treatment once the condition has progressed or pain develops. Consequently, analyzing microbial groups in relation to gingivitis scores could enhance patient outcomes by enabling timely intervention and potentially preventing the progression of chronic periodontitis (Choi et al. 2020; Cai et al. 2021).
In this study, we investigated correlations among 10 periodontal pathogens using the PowerCheck™ Periodontitis Pathogens Multiplex Real-time PCR kit (Socransky et al. 1998; Sim et al. 2024). Due to limited demographic studies focusing specifically on gingivitis, direct comparisons with prior research were constrained. Our study stands out for its novel exploration of specific periodontal pathogens and their relationships with gingivitis within our study population. We observed significant associations between certain microorganisms and the standard gingivitis diagnostic indices such as PI, GI, GR, and BOP. Notably, among these pathogens, F. nucleatum emerged as a potential prognostic marker for gum health, showing associations with these indices independent of age and sex.
F. nucleatum, a member of the oral microbiota, is recognized as a bridging species that connects early colonizers on the tooth surface with late-colonizing pathogens, such as those in the 'Red complex’ (Groeger et al. 2022; Fan et al. 2023). Previous studies have shown that F. nucleatum adheres to and invades oral epithelial cells by binding to host receptors, modulating signaling pathways, and cytokine networks (Groeger et al. 2022; Fan et al. 2023). This process creates an inflammatory environment that disrupts immune homeostasis and can lead to damage of periodontal tissue (Sun et al. 2024). Our study provides clinical evidence that chronic infections with F. nucleatum may contribute to significant gum tissue damage.
This study has several limitations. First, our cross-sectional design and lack of follow-up data did not establish the causality of our findings. The observed associations between F. nucleatum and clinical index measurements for gum highlights the importance of longitudinal studies with follow-up data. Such studies are crucial for gaining a comprehensive understanding of gingivitis and its progression, ultimately leading to more effective prevention and treatment strategies. Second, our focus on 10 periodontal pathogens excludes the broader microbial community, which may also contribute to gingival health. Future studies could explore additional bacterial species to build a more comprehensive understanding of the oral microbiome in gingivitis. Third, we included only patients with gingivitis or mild periodontitis excluding those with chronic periodontitis. Therefore, our findings are confined to the early stages of periodontitis. However, it is worth noting that F. nucleatum has been reported to play a role in both healthy conditions and periodontitis.
CONCLUSION
In conclusion, this study identified associations between ten periodontal pathogens and clinical indicators of early periodontal disease, highlighting the relevance of microbial presence in assessing gingival health. F. nucleatum emerged as significantly associated with gingival indices, suggesting it may contribute to the early inflammatory environment observed in gingivitis and mild periodontitis. Further research incorporating a larger microbial profile and longitudinal designs could enhance our understanding of microbial contributions to periodontal disease, ultimately advancing preventive and therapeutic strategies.
AUTHOR CONTRIBUTIONS
Woo Hyun Shim: Investigation, data curation, writing original draft and editing, and visualization. Jusun Song: data curation, writing original draft and editing, and visualization. Ki Hwan Kim: conceptualization, methodology, writing – review and editing. Sanghun Lee: conceptualization, methodology, investigation, writing – review and editing, project administration, and funding acquisition.
REFERENCES
Aas, J. A., Paster, B. J., Stokes, L. N., Olsen, I., and Dewhirst, F. E. 2005. Defining the normal bacterial flora of the oral cavity. Journal of Clinical Microbiology. 43(11): 5721-5732.
Ali, M. F. and Taha, G. I. 2024. Gingival crevicular fluid volume and protein concentration: A biomarker tool for predicting periodontal diseases progression and severity. Natural and Life Sciences Communications. 23(4): e2024055.
Anderson, G. B., Caffesse, R. G., Nasjleti, C. E., and Smith, B. A. 1991. Correlation of periodontal probe penetration and degree of inflammation. American Journal of Dentistry. 4(4): 177-183.
Armfield, J. M., Slade, G. D., and Spencer, A. J. 2008. Cognitive vulnerability and dental fear. BMC Oral Health. 8(2). https://doi.org/10.1186/1472-6831-8-2
Bartold, P. M. and Van Dyke, T. E. 2019. An appraisal of the role of specific bacteria in the initial pathogenesis of periodontitis. Journal of Clinical Periodontology. 46(1): 6-11.
Cai, Z., Lin, S., Hu, S., and Zhao, L. 2021. Structure and function of oral microbial community in periodontitis based on integrated data. Frontiers in Cellular and Infection Microbiology. 11: 663756.
Calladine, H., Currie, C. C., and Penlington, C. 2022. A survey of patients' concerns about visiting the dentist and how dentists can help. Journal of Oral Rehabilitation. 49(4): 414-421.
Chapple, I. L. C., Mealey, B. L., Van Dyke, T. E., Bartold, P. M., Dommisch, H., Eickholz, P., Geisinger, M. L., Genco, R. J., Glogauer, M., Goldstein, M. et al. 2018. Periodontal health and gingival diseases and conditions on an intact and a reduced periodontium: Consensus report of workgroup 1 of the 2017 world workshop on the classification of periodontal and peri-implant diseases and conditions. Journal of Clinical Periodontology. 45 Supplement 20: S68-S77.
Chattopadhyay, S., Malayil, L., Chopyk, J., Smyth, E., Kulkarni, P., Raspanti, G., Thomas, S. B., Sapkota, A., Mongodin, E. F., and Sapkota, A. R. 2024. Oral microbiome dysbiosis among cigarette smokers and smokeless tobacco users compared to non-users. Scientific Reports. 14(1): 10394.
Checchi, L., Montevecchi, M., Checchi, V., and Zappulla, F. 2009. The relationship between bleeding on probing and subgingival deposits. An endoscopical evaluation. The Open Dentistry Journal. 3: 154-160.
Choi, J. U., Lee, J. B., Kim, K H., Kim, S., Seol, Y. J., Lee, Y. M., and Rhyu, I. C. 2020. Comparison of periodontopathic bacterial profiles of different periodontal disease severity using multiplex real-time polymerase chain reaction. Diagnostics (Basel). 10(11): 965.
Darveau, R. P. 2010. Periodontitis: A polymicrobial disruption of host homeostasis. Nature Reviews Microbiology. 8(7): 481-490.
Elter, J. R., Champagne, C. M., Offenbacher, S., Beck, J. D. 2004. Relationship of periodontal disease and tooth loss to prevalence of coronary heart disease. Journal of Periodontology. 75(6): 782-790.
Fan, Z., Tang, P., Li, C., Yang, Q., Xu, Y., Su, C., and Li, L. 2023. Fusobacterium nucleatum and its associated systemic diseases: Epidemiologic studies and possible mechanisms. Journal of Oral Microbiology. 15(1): 2145729.
Furuta, M., Ekuni, D., Irie, K., Azuma, T., Tomofuji, T., Ogura, T., and Morita, M. 2011. Sex differences in gingivitis relate to interaction of oral health behaviors in young people. Journal of Periodontology. 82(4): 558-565.
Groeger, S., Zhou, Y., Ruf, S., and Meyle, J. 2022. Pathogenic mechanisms of fusobacterium nucleatum on oral epithelial cells. Frontiers in Oral Health. 3: 831607.
Isola, G., Santonocito, S., Lupi, S. M., Polizzi, A., Sclafani, R., Patini, R., and Marchetti, E. 2023. Periodontal health and disease in the context of systemic diseases. Mediators of Inflammation. 2023: 9720947.
Jenkinson, H. F. and Lamont, R. J. 2005. Oral microbial communities in sickness and in health. Trends in Microbiology. 13(12): 589-595.
Kang, M. S., Lee, D. S., Lee, S. A., Kim, M. S., and Nam, S. H. 2020. Effects of probiotic bacterium Weissella cibaria CMU on periodontal health and microbiota: A randomised, double-blind, placebo-controlled trial. BMC Oral Health. 20(1): 243.
Kim, J. and Amar, S. 2006. Periodontal disease and systemic conditions: A bidirectional relationship. Odontology. 94(1): 10-21.
Loe, H. and Silness, J. 1963. Periodontal disease in pregnancy. I. Prevalence and severity. Acta Odontologica Scandinavica. 21: 533-551.
Martinez-Garcia, M. and Hernandez-Lemus, E. 2024. The molecular comorbidity network of periodontal disease. International Journal of Molecular Sciences. 25(18): 10161.
Na, H. S., Kim, S. Y., Han, H., Kim, H. J., Lee, J. Y., Lee, J. H., and Chung, J. 2020. Identification of potential oral microbial biomarkers for the diagnosis of periodontitis. Journal of Clinical Medicine. 9(5): 1549.
Nabila, S., Choi, J., Kim, J. E., Hahn, S., Hwang, I. K., Kim, T. I., Park, H. K., and Choi, J. Y. 2023. Bidirectional associations between periodontal disease and systemic diseases: A nationwide population-based study in Korea. Scientific Reports. 13(1): 14078.
Nazir, M., Al-Ansari, A., Al-Khalifa, K., Alhareky, M., Gaffar, B., and Almas, K. 2020. Global prevalence of periodontal disease and lack of its surveillance. The Scientific World Journal. 2020: 2146160.
Paster, B. J., Boches, S. K., Galvin, J. L., Ericson, R. E., Lau, C. N., Levanos, V. A., Sahasrabudhe, A., and Dewhirst, F. E. 2001. Bacterial diversity in human subgingival plaque. Journal of Bacteriology. 183(12): 3770-3783.
Rahardjo, A., Yoshihara, A., Amarasena, N., Ogawa, H., Nakashima, K., and Miyazaki, H. 2005. Relationship between bleeding on probing and periodontal disease progression in community-dwelling older adults. Journal of Clinical Periodontology. 32(11): 1129-1133.
Scapoli, L., Girardi, A., Palmieri, A., Martinelli, M., Cura, F., Lauritano, D., and Carinci, F. 2015. Quantitative analysis of periodontal pathogens in periodontitis and gingivitis. Journal of Biological Regulators and Homeostatic Agents. 29(3 Suppl 1): 101-110.
Silness, J. and Loe, H. 1964. Periodontal disease in pregnancy. Ii. Correlation between oral hygiene and periodontal condition. Acta Odontologica Scandinavica. 22: 121-135.
Sim, S.-j., Kim, J.-h., and Shin, H.-s. 2024. Preliminary study on the diversity and quantity analysis of oral bacteria according to the sampling methods. Journal of Korean Society of Dental Hygiene. 24(2): 131-139.
Smith, R. G. 1997. Gingival recession. Reappraisal of an enigmatic condition and a new index for monitoring. Journal of Clinical Periodontology. 24(3): 201-205.
Socransky, S. S., Haffajee, A. D., Cugini, M. A., Smith, C., and Kent, R. L. Jr. 1998. Microbial complexes in subgingival plaque. Journal of Clinical Periodontology. 25(2): 134-144.
Sun, J., Feng, S., Ding, T., Wang, T., Du, L., Kang, W., and Ge, S. 2024. Fusobacterium nucleatum dysregulates inflammatory cytokines and NLRP3 inflammasomes in oral cells. Oral Diseases. 30(7): 4767-4781.
Zheng, J., Wang, X., Zhang, T., Jiang, J., and Wu, J. 2024. Comparative characterization of supragingival plaque microbiomes in malocclusion adult female patients undergoing orthodontic treatment with removable aligners or fixed appliances: A descriptive cross-sectional study. Frontiers in Cellular and Infection Microbiology. 14: 1350181.
OPEN access freely available online
Natural and Life Sciences Communications
Chiang Mai University, Thailand. https://cmuj.cmu.ac.th
Supplementary Table 1. Target genes of oral bacteria included in the PowerCheckTM periodontitis pathogens multiplex Real-Time PCR Kit.
|
Socransky complex |
Oral bacteria |
Target gene |
|
Aa complex |
Aggregatibacter actinomycetemcomitans |
fts I |
|
Red complex
|
Porphyromonas gingivalis (P. gingivalis) |
waaA |
|
Tannerella forsythia (T. forsythia) |
fts Z |
|
|
Treponema denticola (T. denticola) |
fts K |
|
|
Orange complex
|
Prevotella intermedia (P. intermedia) |
piACP |
|
Fusobacterium nucleatum (F. nucleatum) |
rpoB |
|
|
Parvimonas micra (P. micra) |
fus A |
|
|
Others
|
Filifactor alocis (F. alocis) |
gyr B |
|
Porphyromonas endodontalis (P. endodontalis) |
16S |
|
|
Treponema socranskii (T. socranskii) |
16S |
Woo Hyun Shim1, Jusun Song2, Ki Hwan Kim1, and Sanghun Lee3, 4, *
1 Department of Dentistry, Myongji Hospital, 55, Hwasu-ro 14, Deogyang-gu, Goyang-si, Gyeonggi-do, Republic of Korea.
2 GC Genome, 107 Ihyeon-ro 30beon-gil, Giheung-gu, Yongin 16924, Gyeonggi-do, Republic of Korea.
3 NH Institute for Natural Product Research, Myongji Hospital, 55, Hwasu-ro 14, Deogyang-gu, Goyang-si, Gyeonggi-do, Republic of Korea.
4 Department of Bioconvergence and Engineering, Graduate school, Dankook University, 152, Jukjeon-ro, Suji-gu, Yongin-si, Gyeonggi-do, Republic of Korea.
Corresponding author: Sanghun Lee, E-mail: shlee92@dankook.ac.kr
Total Article Views
Editor: Anak Iamaroon,
Chiang Mai University, Thailand
Article history:
Received: September 6, 2024;
Revised: November 9, 2024;
Accepted: November 13, 2024;
Online First: November 22, 2024

