
Comparison of Three Genomic DNA Extraction Methods from Sugarcane for Detection of Sugarcane White Leaf Phytoplasma
Supananya Chansri, Parichart Burns, Suriyaphong Nilsang, Wichai Kositratana, and Jutatape Watcharachaiyakup*Published Date : August 23, 2024
DOI : https://doi.org/10.12982/NLSC.2024.052
Journal Issues : Number 4, October-December 2024
Abstract The most suitable DNA extraction method was determined for the efficient detection of sugarcane white leaf phytoplasma (SCWL) in asymptomatic sugarcane. DNA was extracted from the sugarcane midrib samples using three different methods: CTAB, DNeasy® Plant Mini Kit (Qiagen) and DNAsecure Plant Kit (Tiangen). The extracted DNA was evaluated for quantity, quality and downstream application effectiveness. DNA quantity, quality and integrity were determined using A260/280 ratio, A260/230 ratio, gel electrophoresis and qPCR. The CTAB method provided highest yield of 5.65 µg/100 mg fresh tissue followed by DNeasy® Plant Mini Kit (4.53 µg/100 mg) and DNAsecure Plant Kit (1.89 µg/100 mg). A260/280 ratio indicated no significant difference among the three methods. The effectiveness of downstream application was investigated using qPCR amplification of the GADPH housekeeping gene of sugarcane and secY gene were designed in this study, specific to SCWL phytoplasma. The results showed mean Cq values of GADPH and secY were 28.08–30.47 and 33.62–34.69, respectively while CV values were 2.75 and 3.84, respectively with P values less than 0.001. These extraction methods demonstrated effectiveness, highlighting their potential utility in SCWL pathogen detection. The choice of extraction method depended on the primary purpose of the downstream application, user convenience, cost considerations, time efficiency, and accessibility to chemical reagents.
Keywords: SCWL, Detection, Sugarcane, qPCR, secY
Citation: Chansri, S., Burns, P., Nilsang, S., Kositratana, W., and Watcharachaiyakup, J. 2024. Comparison of three genomic DNA extraction methods from sugarcane for detection of sugarcane white leaf phytoplasma. Natural and Life Sciences Communications. 23(4): e2024052.
INTRODUCTION
Sugarcane is an important economic crop in Thailand (Office of the cane and sugar board, 2020). Sugarcane white leaf (SCWL) disease, associated with the presence of SCWL phytoplasma, is one of the most important diseases affecting sugarcane yield (Sroykaew et al., 2018). The disease spreads through infected seedcane and two insect vectors; Matsumuratettix hiroglyphicus and Yamatotettix flavovittatus (Chen, 1978; Hanboonsong et al., 2005). Sugarcane associated with low amount of SCWL phytoplasma may display mild or asymptomatic symptoms (Aljanabi et al., 1999). Consequently, SCWL phytoplasma could rapidly spread to new plantation areas. Therefore, a highly sensitive detection method for SCWL phytoplasma is essential.
During plant DNA extraction, secondary metabolites, lipids, phenolic compounds, and polysaccharides are released from cell lysate. These compounds significantly affect both the quantity and quality of the extracted DNA. They have the potential to interact with nucleic acids and other cellular components (Loomis, 1974). Phenolic compounds, when oxidized, can form covalent bonds with DNA, causing it to turn brown and rendering it unsuitable for research applications (Katterman and Shattuck, 1983). Failure to remove these contaminants can inhibit downstream assays, including PCR (Pedersen et al., 2006; Sarwat et al., 2006; Bellstedt et al., 2010). The method to avoid problems caused by contaminants such as phenolic compounds include freezing the tissue before or during the homogenization process (Katterman and Shattuck, 1983). Many plant species including sugarcane contain high level of polysaccharides and phenolic compounds (Aljanabi et al., 1999). Therefore, it is necessary to obtain an effective method for DNA extraction.
This study aimed to investigate three different DNA extraction methods from the midrib of sugarcane leaf samples contains phytoplasmas associated with SCWL. The quality and quantity of extracted DNA were determined by agarose gel electrophoresis, spectrophotometer, and qPCR amplification.
MATERIALS AND METHODS
Sugarcane sample preparations
Asymptomatic sugarcane leaves contain phytoplasmas associated with SCWL were used in this study. The third leaf from the top was collected from 10 plants, then washed with tap water followed by sterile water, and dried on tissue paper. The midrib of each plant leaf was cut into small pieces (0.2 cm x 0.5 cm) and weighed to obtain 250 mg for three subsamples. Each subsample was ground into a fine powder with liquid nitrogen, then stored at -80°C until the extraction method was performed. Each subsample was further used for each extraction method.
DNA extraction
Cetyltrimethylammonium bromide (CTAB) method
The CTAB method was modified from (Doyle and Doyle, 1987). The resulting sample powder was transferred into a 1.5 mL microtube. Then, 650 µL of CTAB buffer (2% CTAB, 2% PVP, 1.4 M NaCl, 20 mM EDTA, 100 mM Tris-HCl pH 8.0) and 10 µL of 0.2% β – mercaptoethanol were added to the sample. The mixture was incubated for 30 minutes at 65°C. After incubation, plant debris was separated by centrifugation at 12,000 × g for 1 minute and the supernatant was transferred into a new microtube. Six microliters of 50 mg/mL RNase A were added and the mixture was incubated for an hour at 37°C. The solution was then extracted using 650 µL of phenol: chloroform: isoamyl alcohol (PCI) (25:24:1). The extraction was incubated at room temperature for 10 minutes. Subsequently, the aqueous phase was separated by centrifugation at 12,000 x g for 15 minutes and then 200 µL of aqueous solution was transferred into a new microtube. DNA was precipitated by adding 400 µL of ice-cold ethanol and incubated at -20°C for at least 1 hour. The DNA pellet was collected by centrifugation at 12,000 x g for 10 minutes and the supernatant was removed. The pellet was then washed with 500 µL of 70% ethanol and centrifuged at 12,000 x g for 2 minutes. After discarding the supernatant, the pellet was allowed to air dry at room temperature before being resuspended in 50 µL of TE buffer.
DNeasy® plant mini kit (Qiagen) and DNAsecure plant kit (Tiangen)
Sugarcane DNA was extracted following the manufacturer’s protocol for each kit (DNeasy® Plant Mini Kit by Qiagen and DNAsecure Plant Kit by Tiangen) using the same frozen tissue sample (250 mg) as used in the CTAB method.
DNA quality, quantity and integrity analysis
Spectrophotometry
The quantity and quality of DNA were analyzed using spectrophotometry within the absorbance ratios of 260/280 and 260/230 nm with Nabi™ Ultraviolet-Visible (UV/Vis) spectrophotometer (Nano Spectrophotometer, Korea).
Agarose gel electrophoresis
The quality and integrity of DNA were analyzed by agarose gel electrophoresis using 0.8% agarose gel. Electrophoresis was performed using 0.5X Tris–Borate EDTA buffer (40 mM Tris-HCl pH 8.0, 45 mM boric acid, 1 mM EDTA) and a constant voltage of 100 V for 35 minutes. The DNA was stained with 1 μg/mL of ethidium bromide solution. The DNA bands were visualized and photographed under UV – transilluminator (Major Science, Taiwan).
DNA quality investigated by performance in real-time PCR assay (qPCR)
The quality of DNA was analyzed with qPCR amplification of sugarcane glyceraldehyde-3-phosphate dehydrogenase (GAPDH), a housekeeping gene, using specific primers GAPDHF (5’ CAC GGC CAC TGG AAG CA 3’) and GAPDHR (5’ TCC TCA GGG TTC CTG ATG CC 3’) (Iskandar et al., 2004). The experiments for each DNA extraction method were carried out with 10 samples and five replication reactions per sample. The PCR was composed of 0.2 μM of each primer (forward and reverse), 1X Ssofast EvaGreen Supermix (Bio-Rad, USA) and 50 ng DNA template, in a total volume of 20 μL. The qPCR conditions were as follows: an initial cycle of 2 minutes at 98°C, followed by 40 cycles of 5 seconds at 98°C and 5 seconds at 58°C in Bio-Rad CFX96 Touch Real-Time PCR machine (Bio-Rad, USA). The specificity of the amplification product was analyzed through a melt-curve analysis covering a temperature range of 60–95 °C. In addition, qPCR without adding DNA as a non-template control reaction was included to detect any potential false signals originating from DNA contamination or primer dimer formation during the reaction. The analysis was performed using the CFX Maestro Software (Bio-Rad, USA).
Detection of sugarcane white leaf phytoplasma
Primer design
Forward primer and reverse primers were designed to be specific to SCWL phytoplasma based on the secY gene. Nucleotide sequences of SCWL phytoplasma and related species published in GenBank (http://www.ncbi.nlm.nih.gov/) (VWXM01000002.1 NZ_VWOH01000001.1, NZ_JHUK01000003.1, JHUK01000003.1 and NZ_CP025121.1) were analyzed to identify areas specific to SCWL phytoplasma with MEGA 11 (Tamari et al., 2013). Primers were manually designed based on specific regions of secY gene of SCWL phytoplasma. The criteria for designing primers included a length of 18-24 bases, 40–60% G/C content and starting and ending with 1–2 G/C pairs, melting temperature (Tm) of 50–60°C. The primer pairs should have a Tm difference within 5°C and should not contain complementary regions. Each oligonucleotide was analyzed with the Oligonucleotide Properties Calculator program (http://biotools.nubic.northwestern.edu/OligoCalc.html).
Primer specificity was evaluated using samples from ten white leaf grass plants, including Brachiaria mutica (n=2), Zoysia matrella (L.) Merr. (n=3), Cynodon dactylon (n=4), and Ischaemum barbatum (n=1). The presence of phytoplasma associated with white leaf symptom in these sample was confirmed using universal phytoplasma primers (P1: GTCGTAACAAGGTATCCCTACCGG and P2: GGTGGGCCTAAATGGACTTGAACC) as described by Hanboonsong et al., (2006).
qPCR for SCWL phytoplasma detection
The CFX96 Real-Time System (Bio-Rad, USA) was utilized to perform qPCR analyzes on target DNA. The optimum condition of qPCR was analyzed with various annealing temperatures using DNA extracted from asymptomatic sugarcane leaves contain phytoplasmas associated with SCWL. The PCR comprised 1X Ssofast Evergreen Supermix (Bio-Rad, USA), 50 ng template DNA and 0.5 μM of each primer in a total volume of 20 μL. The PCR cycles consisted of an initial cycle of 2 minutes at 98°C, followed by 40 cycles of 5 seconds at 98°C and 5 seconds at 52 – 60 °C (in gradient). The PCR products were evaluated using melt-curve analysis. Furthermore, non-template control was included to identify any spurious signals from DNA contamination or primer dimer formation during the amplification. The qPCR results were analyzed utilizing the CFX Maestro Software (Bio-Rad, USA). The optimized qPCR protocol was employed to assess the quality of 10 samples for each method, with five replication reactions conducted per sample.
Statistical Analysis
The data of DNA quantity, quality and DNA amplification from the three DNA extraction methods were statistically analyzed by a one-way analysis of variance (ANOVA) using Rstudio. Significant difference between each extracted method were determined by Least Significant Difference Test (LSD Test) at P < 0.01.
DNA Quality and Quantity, Estimated Cost, and Time Required
The estimated costs were derived from the prices of chemicals and disposable items, such as pipette tips and microcentrifuge tubes. The cost per sample for each method was calculated in US dollars (USD). Additionally, the time required to complete single extraction procedure for ten sugarcane samples was estimated based on the procedures utilized in this study.
RESULTS
DNA quality, quantity and integrity assessment
Three DNA extraction methods: the CTAB method, the DNeasy® Plant Mini Kit and the DNAsecure Plant Kit produced genomic DNA yield ranging from 14.13 ± 2.519 µg, 11.33 ± 1.654 µg and 4.74 ± 0.461 µg, respectively. The CTAB extraction method had significantly the highest average yield at P < 0.001 (Figure 1A). All three extraction methods produced high quality DNA with minimal protein and phenolic compounds contamination at A260/A280 ratio between 1.80-1.90 without any significant difference (P-value = 0.073) (Figure 1B). However, the A260/A230 ratio showed differences among extraction methods (P-value = <0.001). The second measure for DNA purity was A260/A230 ratio with acceptable range of high DNA purity between 2.0-2.2 (Lucena-Aguilar et al., 2016). The genomic DNA extracted using the CTAB method exhibited an A260/A230 ratio ranging from 1.71-2.03, indicating the presence of a small amount of contaminants of phenol and protein. These contaminants may have originated from both the CTAB extraction method and cellular components. The DNeasy® Plant Mini Kit and DNAsecure Plant Kit produced the DNA with higher ranges of ratio from 2.51-2.76 and 2.13-3.67, respectively (Figure 1C). This suggests that the DNA was contaminated with substances such as carbohydrates and salts.
Visual inspection of gel electrophoresis showed that the extracted DNA using the DNeasy® Plant Mini Kit and DNAsecure Plant Kit exhibited better quality than the DNA obtained through the CTAB method. The latter method produced high molecular weight DNA (>20 kb) smear (HMW smear) (Lucena-Aguilar et al., 2016) (Figure 2).

Figure 1. Box plot diagram of DNA concentration (A) absorbance ratio of A260/ A280 (B) and A260/A230 (C) obtained for each extraction method (CTAB, DNeasy@ Plant Mini Kit: DNeasy and DNAsecure Plant Kit: DNAsecure). Each box indicates the values of the quartiles, the limits of the distribution represent the maximum and minimum values. Different letters indicate significant differences P < 0.01) among the evaluated methods according to the Least Significant difference.
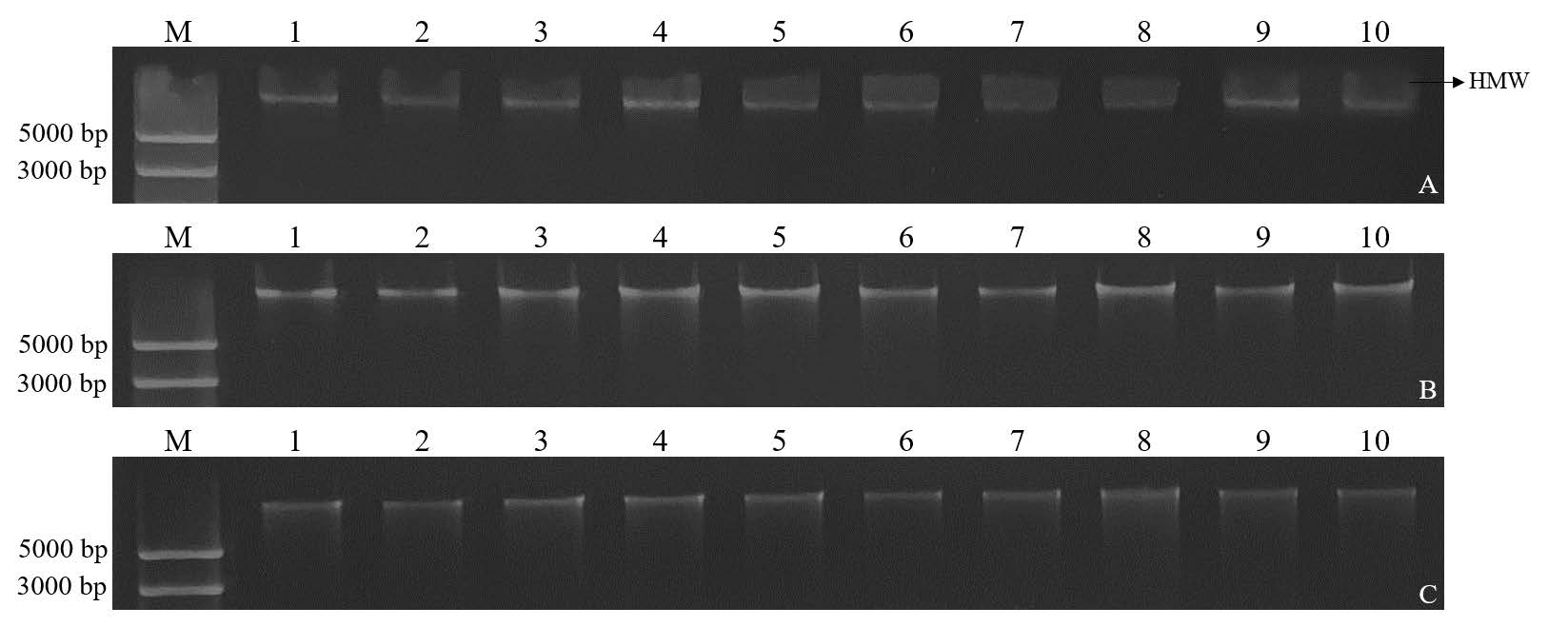
Figure 2. Agarose gel electrophoresis of total genomic DNA (lane 1 – 10) extracted from sugarcane by using the three different extraction methods: CTAB method (A) DNeasy® Plant Mini Kit (B) and DNAsecure Plant Kit (C). Lane M; O'GeneRuler Express DNA Ladder (Thermo Scientific™), HMW: high molecular weight
DNA quality investigated by amplification efficiency in qPCR
The DNeasy® Plant Mini Kit showed the highest efficiency in sugarcane GAPDH amplification, reflected by the lowest mean Cq value (28.08 ± 0.08), followed by the CTAB method (29.59 ± 0.18) and DNAsecure Plant Kit (30.47 ± 0.16) (Table 2). Interestingly, despite a statistically significant difference (P < 0.001), the mean Cq values differed by only 1-2 units between the methods. All three extraction methods displayed high efficiency of all repeatability and consistent results with low variability of CV 2.75% across all methods.
Table 1. The mean ± SD values of Cq were obtained from qPCR amplification of sugarcane GAPDH using DNA extracted by three different methods from asymptomatic sugarcane leaves contain phytoplasmas associated with SCWL.
|
DNA extraction methods |
Cq (mean) ± SD1 |
|
CTAB |
29.59 ± 0.18 b |
|
DNeasy® Plant Mini Kit |
28.08 ± 0.08 a |
|
DNAsecure Plant Kit |
30.47 ± 0.16 c |
|
CV |
2.75 |
|
P-value |
<0.001 |
Note: 1Mean followed by different lowercase letter are significantly different at P< 0.01 (one way ANOVA and LSD test).
Detection of phytoplasma associated with sugarcane white leaf
A qPCR assay was developed to detect phytoplasma associated with SCWL by designing primers targeting the secY gene based on sequence comparison of multiple phytoplasmas strains, with the detailed primer sequences provided in Table 2. Optimal conditions were determined by testing various annealing temperatures, with 60°C proving to be the most effective, as evidenced by a single peak in melt peak analysis (Figure 3A and 3B). Subsequently, the qPCR product was analyzed through agarose gel electrophoresis, confirming the presence of a single target band of the expected product size (results not shown). Notably, this condition exhibited no amplification with non-target phytoplasmas (Figure 4A and 4B). These results demonstrate the high specificity of these primers.
Table 2. Primer sequences specific to phytoplasma associated with SCWL targeted on secY gene were designed in this study.
|
Name |
Sequence |
Source |
|
secYI-F |
5' AGG AAG TTC ACG TCC TTT ATG 3' |
this study |
|
secYI-R |
5' TAC AAT AGA AGC AGT TAT GTA AGG 3' |
this study |
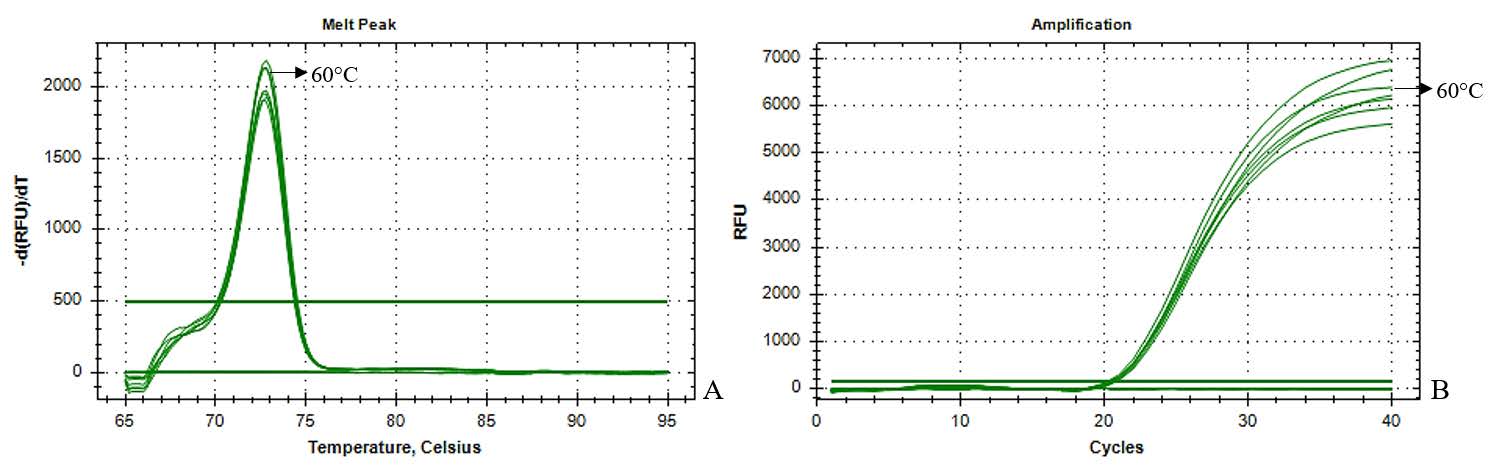
Figure 3. qPCR amplification with secY1-F and secY1-R primers with various annealing temperatures (52-60°C) (A) Melt peak analysis and (B) the amplification plot of qPCR product amplified.
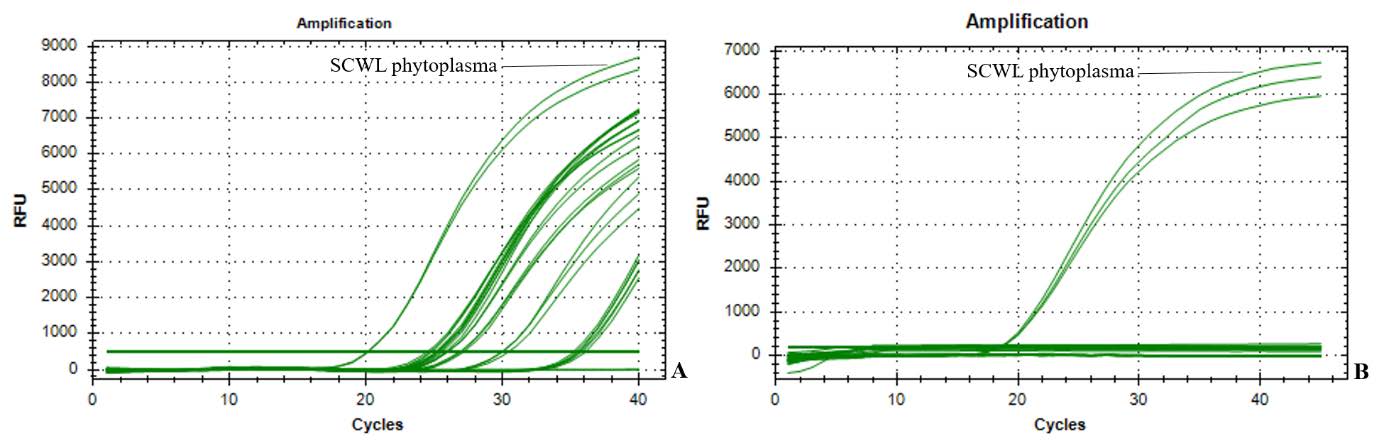
Figure 4. The amplification plot of qPCR product amplified with 10 white leaf glass samples and sugarcane white leaf sample (A) the results showed the successful amplification of phytoplasma DNA with universal P1/P2 primers, thus confirming the phytoplasma associated in all tested samples. (B) The qPCR amplification with secY1-F/secY1-R primers indicated the specificity of these primer to phytoplasma associated with sugarcane white leaf (SCWL phytoplasma).
The effectiveness of three DNA extraction methods was investigated through qPCR analysis of phytoplasma associated SCWL in asymptomatic sugarcane midrib samples. The DNeasy® Plant Mini Kit yielded the lowest mean Cq value (33.62 ± 0.91), reflecting the highest effectiveness of detection. CTAB (34.18 ± 1.53) and DNAsecure Plant Kit (34.69 ± 1.41) produced slightly higher Cq values, indicating lower effectiveness of detection. Although statistically significant (P < 0.001), the Cq value differences were relatively small. All three methods displayed reliable detection across replicates of positive results in all 10 samples with low variability of CV at 3.84 across all methods (Table 3).
Table 3. The mean ± SD values of Cq were obtained from qPCR amplification of the secY using DNA extracted by three different methods from asymptomatic sugarcane leaves contain phytoplasmas associated with SCWL.
|
DNA extraction methods |
Cq (mean) ± SD1 |
|
CTAB |
34.18 ± 1.53a |
|
DNeasy® Plant Mini Kit |
33.62 ± 0.91b |
|
DNAsecure Plant Kit |
34.69 ± 1.41a |
|
CV |
3.841 |
|
P |
<0.001 |
Note: 1 Mean followed by different lowercase letter are significantly different at P< 0.01 (one way ANOVA and LSD test).
In conclusion, this study evaluated three DNA extraction methods: CTAB, DNeasy® Plant Mini Kit, and DNAsecure Plant Kit, each tested with 10 samples. The CTAB method yielded high amounts of DNA with only acceptable quality and qPCR performance and is priced at $24. However, it requires a high level of skill and is time-consuming, taking about 6 hours to complete. The DNeasy® Plant Mini Kit provided very satisfactory DNA quality and qPCR results with a shorter processing time of 2.5 hours, but it is the most expensive at $70. The DNAsecure Plant Kit, while the cheapest at $17 and fastest at 2.5 hours, produced lower DNA yields with only acceptable quality and qPCR performance (Table 4). All extraction methods required the same general molecular laboratory equipment, except that the CTAB method requires additional fume hood use.
Table 4. Comparison DNA yield, quality, integrity, cost and time of three extraction methods from asymptomatic sugarcane leaves contain phytoplasmas associated with SCWL.
|
Evaluation |
CTAB (n = 10) |
DNeasy® Plant Mini Kit (n = 10) |
DNAsecure Plant Kit (n = 10) |
|
|
DNA yield rate |
+++ |
++ |
+ |
|
|
Absorbance ratio A260/280 |
+++ |
+++ |
+++ |
|
|
Absorbance ratio A260/230 |
++ |
++ |
++ |
|
|
HMW smear |
++ |
+++ |
+++ |
|
|
Skill requirement |
High |
Moderate |
Moderate |
|
|
qPCR |
++ |
+++ |
++ |
|
|
Cost (USD)1 |
24 |
70 |
17 |
|
|
Time-consuming (hour) |
6 |
2.5 |
2.5 |
|
Note: +++, very satisfactory; ++, acceptable; +, questionable. HMW, high molecular weight (>20 kb) DNA. 1Cost was calculated based on chemical and consumable expenses, with an exchange rate of 1 USD = 36 THB. All costs are based on recent catalog pricing (July 2024).
DISCUSSION
DNA quality and integrity is an important factor influencing the effectiveness of downstream applications (Fornasier et al., 2014). The DNA extraction method serves as the initial and pivotal step in most molecular research (Allen et al., 2006; Dittrich-Schröder et al., 2012).
Sugarcane is a plant species that produces and accumulates several substances including starch, sugars and phenolic compounds that could interfere with DNA extraction and downstream applications (Bermúdez et al., 2016). Though DNA extraction methods from sugarcane were previously reported (Bermúdez et al., 2016; Esfandani-Bozchaloyi et al., 2019), there was little studies on DNA yield, quality, and integrity. The CTAB method exhibited high DNA yield from sugarcane with minute contamination of chemicals such as phenol. The CTAB method is most used for DNA extraction especially high polysaccharide plant species such as corn, rice, wheat, yam, taro, cassava and berry plants (Webb and Knapp, 1990; Sharma et al., 2010). CTAB functions as a cell lysis reagent and at high ionic strength solution, forms a complex with the polysaccharides, subsequently removed in the next PCI extraction step (Vinod, 2004). The PCI aids in achieving high purity DNA by eliminating protein and polysaccharide contaminants (Greco et al., 2014). However, incomplete removal of these substances may inhibit the PCR process (Tamari et al., 2013). In our case, the slight lower A260/230 ratio indicated the minute amount of these chemicals and lower DNA integrity with high molecular weight DNA (HMW) smear. Proficiency in this critical step significantly impacts DNA purity, influencing subsequent steps (Pedersen et al., 2006; Sarwat et al., 2006; Bellstedt et al., 2010). The high yield from the CTAB method was similar to previously reported (Sarwat et al., 2006). On the other hand, the extraction methods using commercial kits, offered streamline extraction with relative time saving. However, high costs, lower yield, and occasional non-repeatability of DNA yields in certain cases have been reported (Akkurt, 2012; Silva and Domingues, 2015). In addition, commercial extraction kits yielded an amplification ratio of 30-60%, while the traditional method achieved 70-100% (Akkurt, 2012).
The usability of DNA from three extraction methods for downstream applications was determined by qPCR assay for the sugarcane GAPDH and detection of phytoplasma associated SCWL using secY amplification. While all DNA samples were amplified of both genes, the Cq value from extract DNA by DNeasy® Plant Mini Kit was significantly lowest by 1-2 cycles. The CTAB method was the second most effective method. This study yielded comparable DNA quality for qPCR with a narrow range of Ct values and consistent results. Similarly, three DNA extraction methods for Peganum harmala, Tamarix ramosissima, and Potentilla reptans resulted in comparable DNA quality and consistent PCR amplification percentages (Salehi et al., 2023).
In addition, the DNA extraction method should be efficient, cost-effective, require less time and labor (Lagisz et al., 2010). This study found that the DNeasy® Plant Mini Kit provided the best extraction method, although it was the most expensive. However, since the primary purpose of the downstream application was for high accuracy detection, cost was not a major concern. This method may not be suitable for large-scale applications, but it is useful for screening seedcane for SCWL disease before tissue culture propagation. If high accuracy detection is required on a large scale, the higher cost may be justified despite the expense.
CONCLUSION
Both the manual CTAB DNA extraction method and two commercial kit DNA extraction methods yielded DNA of sufficient quantity and quality for detecting the phytoplasma associated SCWL in sugarcane midrib sample. However, optimal method selection should consider user preferences and constraints, including cost, time-consuming, skill requirement, resource availability, equipment access, and the ability to handle large sample sets. Cost is a crucial factor and should be evaluated carefully, especially in large-scale studies where budget constraints can significantly impact the choice of extraction technique. Additionally, the downstream application and purpose of the extraction can sometimes be more important than cost, such as in the case of detection accuracy. For applications where sensitivity and high-throughput detection of phytoplasma associated with SCWL are prioritized, commercial kits with superior sensitivity might be preferred, depending on these factors.
ACKNOWLEDGEMENTS
The laboratory facilities and funding were supported by Center of Excellence on Agricultural Biotechnology: (AG-BIO/MHESI), Bangkok 10900, Thailand and Center for Agricultural Biotechnology, Kasetsart University under the project Development of droplet digital PCR (ddCPR) for detection of sugarcane white leaf phytoplasma” project no. CAB-KU/66-001-003.
AUTHOR CONTRIBUTIONS
Supananya Chansri, Jutatape Watcharachaiyakup, Wichai Kositratana and Parichart Burns designed and conducted all the experiments and also contributed to writing the manuscript. Suriyaphong Nilsang performed statistical analysis and data visualization. All authors have read and approved the final manuscript.
CONFLICT OF INTEREST
The authors declare that they hold no competing interests.
REFERENCES
Akkurt, M. 2012. Comparison between modified DNA extraction protocols and commercial isolation kits in grapevine (Vitis vinifera L.). Genetics and Molecular Research. 11: 2343-2351.
Aljanabi, S. M., Forget, L., and Dookun, A. 1999. An improved and rapid protocol for the isolation of polysaccharide-and polyphenol-free sugarcane DNA. Plant Molecular Biology Reporter. 17: 281-282.
Allen, G. C., Flores-Vergara, M., Krasynanski, S., Kumar, S., and Thompson, W. 2006. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nature Protocols. 1: 2320-2325.
Bellstedt, D. U., Pirie, M. D., Visser, J. C., de Villiers, M. J., and Gehrke, B. 2010. A rapid and inexpensive method for the direct PCR amplification of DNA from plants. American Journal of Botany. 97: e65-e68.
Bermúdez, M., Guzmán, S., Orozco, M., Velázquez, J., Buenrostro, M., and Michel, C. 2016. Optimizing a protocol for DNA isolation of leaf Saccharum officinarum. Revista Mexicana de Ciencias Agrícolas. 7: 897-910.
Chen, C. 1978. Vector-pathogen relationships of sugarcane white leaf disease. Taiwan Sugar. 25: 50-54.
Dittrich-Schröder, G., Wingfield, M. J., Klein, H., and Slippers, B. 2012. DNA extraction techniques for DNA barcoding of minute gall-inhabiting wasps. Molecular Ecology Resources. 12: 109-115.
Doyle, J. J., and Doyle, J. L. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin. 19: 11-15.
Esfandani-Bozchaloyi, S., Sheidai, M., and Kalalegh, M. H. 2019. Comparison of DNA extraction methods from Geranium (Geraniaceae). Acta Botanica Hungarica. 61: 251-266.
Fornasier, F., Ascher, J., Ceccherini, M., Tomat, E., and Pietramellara, G. 2014. A simplified rapid, low-cost and versatile DNA-based assessment of soil microbial biomass. Ecological Indicators. 45: 75-82.
Greco, M., Saez, C. A., Brown, M. T., and Bitonti, M. B. 2014. A simple and effective method for high quality co-extraction of genomic DNA and total RNA from low biomass Ectocarpus siliculosus, the model brown alga. PloS One. 9: e96470.
Hanboonsong, Y., Ritthison, W., and Chosai, C. 2005. Molecular detection and transmission of sugarcane white leaf Phytoplasma in leafhoppers (Thai). Asia-Pacific Journal of Science and Technology. 10: 13-21.
Iskandar, H. M., Simpson, R. S., Casu, R. E., Bonnett, G. D., Maclean, D. J., and Manners, J. M. 2004. Comparison of reference genes for quantitative real-time polymerase chain reaction analysis of gene expression in sugarcane. Plant Molecular Biology Reporter. 22: 325-337.
Katterman, F., and Shattuck, V. 1983. An effective method of DNA isolation from the mature leaves of Gossypium species that contain large amounts of phenolic terpenoids and tannins. Preparative Biochemistry. 13: 347-359.
Lagisz, M., Port, G., and Wolff, K. 2010. A cost‐effective, simple and high‐throughput method for DNA extraction from insects. Insect Science. 17: 465-470.
Loomis, W. 1974. Overcoming problems of phenolics and quinones in the isolation of plant enzymes and organelles. Methods in Enzymology. 31:528-544.
Lucena-Aguilar, G., Sánchez-López, A. M., Barberán-Aceituno, C., Carrillo-Avila, J. A., López-Guerrero, J. A., and Aguilar-Quesada, R. 2016. DNA source selection for downstream applications based on DNA quality indicators analysis. Biopreservation and Biobanking. 14: 264-270.
Office of the cane and sugar board. 2020. Sugarcane Cultivation Situation Report 2019/2020. Retrieved 20 Febuary 2024 from http://www.ocsb.go.th/upload/journal/fileupload/13813-5872.pdf
Pedersen, N., Russell, S. J., Newton, A. E., and Ansell, S. W. 2006. A novel molecular protocol for the rapid extraction of DNA from bryophytes and the utility of direct amplification of DNA from a single dwarf male. The Bryologist. 109: 257-264.
Salehi, Z., Amirahmadi, A., Rezaei, A., Farrokh, P., and Ghasemian, J. 2023. Comparison of five DNA extraction methods in three medicinal plants: Peganum harmala L., Tamarix ramosissima Ledeb., and Potentilla reptans L. Molecular Biology Research Communications. 12: 1.
Sarwat, M., Singh Negi, M., Lakshmikumaran, M., Kumar Tyagi, A., Das, S., and Shankar Srivastava, P. 2006. A standardized protocol for genomic DNA isolation from Terminalia arjuna for genetic diversity analysis. Electronic Journal of Biotechnology. 9(1): 86-91.
Sharma, P., Joshi, N., and Sharma, A. 2010. Isolation of genomic DNA from medicinal plants without liquid nitrogen. Indian Journal of Experimental Biology. 48: 610–614.
Silva, D. M., and Domingues, L. 2015. On the track for an efficient detection of Escherichia coli in water: A review on PCR-based methods. Ecotoxicology and Environmental Safety. 113: 400-411.
Sroykaew, J., Leksakul, K., Boonyawan, D., and Akarapisan, A. 2018. Treating white leaf disease in sugarcane cuttings using solution plasma. Natural and Life Sciences Communications. 17: 61-72.
Tamari, F., Hinkley, C. S., and Ramprashad, N. 2013. A comparison of DNA extraction methods using Petunia hybrida tissues. Journal of Biomolecular Techniques. 24: 113.
Vinod, K. K. 2004. Total genomic DNA extraction, quality check and quantitation. p.109-121. In: Proceedings of the training programme on "Classical and modern plant breeding techniques a hands on training", 1-21 Nov 2004. Tamil Nadu Agricultural University, Coimbatore.
Webb, D. M., and Knapp, S. J. 1990. DNA extraction from a previously recalcitrant plant genus. Plant Molecular Biology Reporter. 8: 180-185.
OPEN access freely available online
Natural and Life Sciences Communications
Chiang Mai University, Thailand. https://cmuj.cmu.ac.th
Supananya Chansri1, 2, Parichart Burns3, Suriyaphong Nilsang4, Wichai Kositratana1, 2, 5, and Jutatape Watcharachaiyakup1, 2, *
1 Center for Agricultural Biotechnology, Kasetsart University, Kamphaeng Saen Campus, Nakhon Pathom 73140, Thailand.
2 Center of Excellence on Agricultural Biotechnology: (AG-BIO/MHESI), Bangkok 10900, Thailand.
3 National Center for Genetic Engineering and Biotechnology, National Science and Technology Development Agency, Pathum Thani 12120, Thailand.
4 Department of Production Engineering, Faculty of Engineering, King Mongkut’s University of Technology Thonburi (KMUTT), Bangkok 10140 Thailand.
5 Department of Plant Pathology, Faculty of Agriculture at Kamphaeng Saen, Kasetsart University, Nakhon Pathom, 73140, Thailand
Corresponding author: Jutatape Watcharachaiyakup, E-mail: jutatape.w@ku.th
Total Article Views
Editor: Tonapha Pusadee,
Chiang Mai University, Thailand
Article history:
Received: February 26, 2024;
Revised: July 20, 2024;
Accepted: August 1, 2024;
Online First: August 23, 2024

