
Identification of Early Protein Damage-Associated Molecular Pattern (DAMPs) Candidates from Cardiac Cells Subjected to an In Vitro Myocardial Ischemic Injury
Sarawut Kumphune, Nitchawat Paiyabhroma, Porrnthanate Seenak, Worawat Songjang, Arunya Jiraviriyakul, Noppadon Jumroon, Panyupa Pankhong, Siriwan Thaisakun, Narumon Phaonakrop, Sittiruk Roytrakul , and Nitirut Nernpermpisooth*Published Date : July 3, 2024
DOI : https://doi.org/10.12982/NLSC.2024.047
Journal Issues : Number 3, July-September 2024
Abstract Damage-associated molecular patterns (DAMPs) are intracellular molecules that are released from cells that undergo injury. Protein-DAMPs are abundant biomolecules that could potentially be a novel biomarker for pathology. Myocardial ischemia injury is a causative process for ischemic heart disease, which is now the leading cause of global deaths. Early diagnosis is necessary to save patient lives, so the discovery of new early cardiac biomarkers is crucial. Here, we examined the protein-DAMPs expression in ischemic buffer collected from adult ventricular myocytes cells subjected to various timepoints of simulated ischemia (sI). Adult ventricular myocytes cells (AC-16) were subjected to sublethal simulated ischemia (sI) by treatment with ischemic buffer for 5, 10, 20, and 30 minutes (I5, I10, I20, and I30, respectively). After simulated ischemia was achieved, the ischemic buffer was collected and determined for proteins profiling by Shotgun proteomics approach. Cell viability was reduced in a time dependent manner in ischemia. Targeted proteomics reveals 21 proteins found to highly increase in early ischemic injury and can still be detected until 30 minutes of ischemia. Lactotransferrin (LTF) was determined to be the protein with the greatest impact, exhibiting both fast and sustained release throughout the whole study time. The present study identified the protein damage associated molecular pattern (DAMPs) released specifically from ischemia-induced cardiomyocytes injury, which could potentially be candidates to be early biomarkers for myocardial ischemic injury. Further studies should be focused on in vivo validation of potential protein markers that could be used in practical clinical applications.
Keywords: Ischemic heart diseases, Ischemia, Damage-associated molecular patterns, DAMPs, Early biomarkers, Proteomics
Funding: The present study was supported by Naresuan University and National Science, Research, and Innovation Fund (NSRF) (grant number 65A107000032, R2565B088) for N.N., and A.J. This study was partially supported by Chiang Mai University, Thailand through Biomedical Engineering and Innovation Research Center, Chiang Mai University, Thailand.
Citation: Kumphune, S., Paiyabhroma, N., Seenak, P., Songjang, W., Jiraviriyakul, A., Jumroon, N., Pankhong, P., Thaisakun, S., Phaonakrop, N., Roytrakul, S., and Nernpermpisooth, N. 2024. Identification of early protein damage-associated molecular pattern (DAMPs) candidates from cardiac cells subjected to an in vitro myocardial ischemic injury. Natural and Life Sciences Communications. 23(3): e2024047.
INTRODUCTION
Despite advancements in health knowledge and technology, myocardial ischemia continues to be the primary cause of mortality worldwide and is expected to remain so in the near future (Mozaffarian et al., 2016). In most cases, a myocardial infarction is caused by a coronary occlusion, which cuts off blood flow to the heart and causes permanent death (Jennings and Reimer, 1991). During myocardial ischemia and reperfusion, reactive oxygen species (ROS) are produced alongside underlying mechanisms including cardiac cell apoptosis (Yellon and Hausenloy, 2007).
Clinical examination and biochemical marker alterations diagnose acute myocardial infarction (AMI) (Johanson et al., 2003). One of the best ways to diagnose myocardial damage is to measure cardiac biomarkers. Conventional cardiac indicators like CK-MB and cTnT/cTnI are sensitive and specific diagnostics for myocardial necrosis. They rise more 3–6 hours after myocardial cell damage, therefore patients may have to wait to be detected and treated (Charpentier et al., 2010). Conventional biomarkers may not increase during reversible myocardial ischemia, and additional diagnostic methods such as stress testing and echocardiography are not regularly accessible in the emergency department (Charpentier et al., 2010). Thus, the identification of early cardiac biomarkers, which reflect the early phase of ischemia damage, aids in faster diagnosis and better medical therapy.
In response to cellular damage and death, cellular components are released into the extracellular space and circulation, where they function as damage-associated molecular patterns (DAMPs) (Land, 2015; Roh and Sohn, 2018). The well-known cellular DAMPs include cellular proteins such as Heat Shock Proteins (HSPs), High Mobility Group Box-1 (HMGB-1), Adenosine Triphosphate (ATP), nuclear and mitochondrial DNA (mtDNA), and RNA) (Land, 2015; Roh and Sohn, 2018). Several DAMPs have been identified from experimental MI models in an effort to replicate clinical ischemia/reperfusion injury as closely as feasible and to investigate possible DAMP-related therapies. The significance of cardiac DAMPs as a diagnostic molecule that could potentially serve as an early cardiac biomarker has been less thoroughly studied. In addition, the majority of reported cardiac DAMPs for myocardial infarction originated from ischemia and reperfusion injury (Roh and Sohn, 2018) and consequently less information on ischemic cardiac DAMPs has been reported. In this current study, we identify cardiac specific protein-DAMPs in response to in vitro ischemic injury by the proteomic approach. The findings from this study provide fundamental evidence for further investigation in pre-clinical in vivo animal model of acute myocardial infarction (AMI), and validation of clinical applications of novel biomarkers.
MATERIALS AND METHODS
Materials
All basic chemicals were purchased from Sigma (Sigma, St. Louis, MO, USA). Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum and trypsin-EDTA were purchased from Gibco (Gibco-Thermo Fisher Scientific, USA). CyQUANT™ LDH Cytotoxicity Assay Kit, and 3-(4,5-dimethyl-2-thiazol)-2,5-diphenyl-2Htetrazolium bromide (MTT) were purchased from Invitrogen (Invitrogen-Thermo Fisher Scientific, USA).
Cell type and cell culture
Ventricular myocytes cell line (AC16) was purchased from American Type Cell Culture (ATCC-CRL-3568) and was cultured in Dulbecco's Modified Eagle Medium/Nutrient Mixture F-12 (DMEM/F12) supplemented with 10% fetal Bovine Serum (FBS), 5,000 units of penicillin and streptomycin. Cells were cultured at 37°C, 5% CO2 + 95% Air throughout the experiments.
Simulated ischemia (sI) protocol
Cells were washed once with PBS before addition of 2 ml of ischemic buffer or control buffer. Sublethal simulated ischemia was induced by treatment of myocytes for 5, 10, 20, and 30 minutes (I5, I10, I20, and I30, respectively) at 37°C with a modified Krebs buffer (ischemic buffer) as previously published (Nernpermpisooth et al., 2017). The control group was treated with Krebs buffer supplemented with 20mM D-glucose and 1mM sodium pyruvate (control buffer) for 5, 10, 20, and 30 minutes. After simulated ischemia was achieved, the ischemic buffer was collected for further analysis as ischemic samples. The control buffer of every timepoint was pooled up and used as a control group. The experiment was performed in duplicate in three independent experiments (n = 6/conditions).
Determination of cell viability and cellular injury
The measurement of cell viability was performed by MTT cell survival assay. The percentage of cell viability was calculated by comparing the optical density of treated samples with untreated control group (100% viability). The released-lactate dehydrogenase (LDH) activity was measured to indicate the loss of permeability of the cell membrane. The LDH activity, in the cell culture medium, was measured by The CyQUANT LDH Cytotoxicity Assay Kit.
Determination of intracellular reactive oxygen species (ROS) production
A 1 × 105 cells/ml of cells was cultured in a 96-well black plate for 24 h. Prior to treat with ROS fluorescence indicator, the culture media was removed, and the cells washed once with PBS before incubating with complete media containing 250 μM carboxy-H2DCFDA for 30 minutes. Then cells were subjected to sI for indicated times. At the end of sI, the fluorescence intensity was measured for determination the intracellular ROS level by using microplate reader Tecan Infinite® 200 PRO (Tecan Trading AG, Switzerland) with excitation wavelength of λ 498 nm and emission wavelength of λ 522 nm (Paiyabhroma, 2018).
Cellular protein collection and determination of protein concentration
After finishing the treated conditions, the cells were lysed in HEPES pH 8, 0.5% SDS and a protease inhibitor cocktail. The protein concentration was determined by Bradford assay for protein carbonyl level measured at absorbance of 595 nm.
Protein separation by SDS-polyacrylamide gel electrophoresis
Protein samples were separated into 12% acrylamide (29:1 acrylamide: bis-acrylamide). The samples, which mixed with sample buffer, were heated at 95°C for 5 minutes and electrophoresis at a constant 200 V until the bromophenol blue front dye reached the bottom of the gel. The gels were washed off the glass plates and washed twice with deionized water, followed by fixing with acid alcohol fixative solution for 1 hour. Then, the gels were stained with Coomassie R-250 dye at room temperature for 3 hours with shaking. For destaining, gels were shaking with destaining solution (50% (v/v) methanol in water with 10% (v/v) acetic acid) until the protein bands could be observed without background staining of the gel.
Sample preparation for shotgun proteomics
The ischemic buffer or reperfusion medium was transferred to a new tube, mixed well with 2 volumes of cold acetone, and incubated overnight at -20°C. The mixture was centrifuged at 10,000 x g for 15 minutes and the supernatant was discarded. The pellet was dried and stored at -80°C prior to use.
In solution trypsin digestion
Protein concentration of collected samples was determined by Lowry assay using BSA as a standard protein (Lowry et al., 1951). Five micrograms of protein samples were subjected to in-solution digestion. Samples were completely dissolved in 10 mM ammonium bicarbonate (AMBIC), reduced disulfide bonds using 5 mM dithiothreitol (DTT) in 10 mM AMBIC at 60°C for 1 hour and alkylation of sulfhydryl groups by using 15 mM Iodoacetamide (IAA) in 10 mM AMBIC at room temperature for 45 minutes in the dark. The protein samples were digested with sequencing grade porcine trypsin (1:20 ratio) for 16 h at 37°C. The tryptic peptides were dried using a speed vacuum concentrator and resuspended in 0.1% formic acid for nano-liquid chromatography tandem mass spectrometry (nanoLC-MS/MS) analysis.
Liquid chromatography-tandem mass spectrometry (LC/MS-MS)
The tryptic peptide samples were prepared for injection into an Ultimate3000 Nano/Capillary LC System (Thermo Scientific, UK) coupled to a ZenoTOF 7600 mass spectrometer (SCIEX, Framingham, MA, USA). Briefly, one microliter of peptide digests was enriched on a µ-Precolumn 300 µm i.d. X 5 mm C18 Pepmap 100, 5 µm, 100 A (Thermo Scientific, UK), separated on a 75 μm I.D. x 15 cm and packed with Acclaim PepMap RSLC C18, 2 μm, 100Å, nanoViper (Thermo Scientific, UK). The C18 column was enclosed in a thermostatted column oven set to 60°C. Solvent A and B containing 0.1% formic acid in water and 0.1 % formic acid in 80% acetonitrile, respectively were supplied on the analytical column. A gradient of 5–55% solvent B was used to elute the peptides at a constant flow rate of 0.30 μl/minute for 30 minutes.
The source and gas parameters on the ZenoTOF 7600 system for all acquisitions were as follows: the ion source gas 1 was set to 8 psi, the curtain gas was set to 35 psi, the CAD gas was set to 7 psi, the source temperature was set to 200°C, the polarity was set to positive, and the spray voltage was set to 3300 V.
DDA method selection of the top, most abundant top 50 precursor ions per survey MS1 for MS/MS at an intensity threshold exceeding 150 cps. Sampled precursor ions were dynamically excluded for 12 s after two incidences of MS/MS sampling occurrence (MS/MS sampling with dynamic CE for MS/MS enabled). The MS2 spectra were collected from 100–1,800 m/z with a 50-ms accumulation time and Zeno trap enabled. For the collision energy equation, the declustering potential was set to 80 V, with 0 V DP spread, and the CE spread was set to 0 V. The time bins to sum were set to 8 with all channels enabled and a 150,000 cps Zeno trap threshold. The cycle time for the Top 60 DDA method was 3.0 s.
Bioinformatics and data analysis
MaxQuant 2.2.0.0 was used to quantify the proteins in individual samples using the Andromeda search engine to correlate MS/MS spectra to the Uniprot Homo sapiens database (Tyanova, Temu, and Cox, 2016). Label-free quantitation with MaxQuant's standard settings was performed: maximum of two miss cleavages, mass tolerance of 0.6 Dalton for main search, trypsin as digesting enzyme, carbamidomethylation of cysteine as fixed modification, and the oxidation of methionine and acetylation of the protein N-terminus as variable modifications. Only peptides with a minimum of 7 amino acids, as well as at least one unique peptide, were required for protein identification. Only proteins with at least two peptides, and at least one unique peptide, were considered as being identified and used for further data analysis. Protein FDR was set at 1% and estimated by using the reversed search sequences. The maximal number of modifications per peptide was set to 5. As a search FASTA file, the proteins present in the Homo sapiens proteome downloaded from Uniprot was performed further analysis. Potential contaminants present in the contaminants, fast file that comes with Macquet were automatically added to the search space by the software.
Data analysis
The Macquet ProteinGroups.txt file was loaded into Perseus version 1.6.6.0 (Tyanova, Temu, et al., 2016), potential contaminants that did not correspond to any UPS1 protein were removed from the data set. Max intensities were log2 transformed and p comparisons between conditions were done via One-way ANOVA. Missing values were also imputed in Perseus using constant value (zero). The visualization and statistical analyses were conducted using the MultiExperiment Viewer (MeV) in the TM4 suite software, Grapbio1 (Zhao and Wang, 2022) and Metaboanalyst (Howe et al., 2011; Pang et al., 2022). Protein organization and biological action was investigated conforming to protein analysis through evolutionary relationships (Panther) protein classification (Mi et al., 2019). Venn diagrams was used to show the differences between protein lists originating from different differential analyses (Bardou et al., 2014). The STITCH database version 5 was used to analyze the common and the forecasted functional interaction networks between identified proteins and small molecules (Szklarczyk et al., 2016).
Determination of lactoferrin by ELISA
Lactoferrin concentration was determined from collected samples by Human Lactoferrin ELISA Kit (Abcam, Cambridge, UK) according to the manufacturer's instructions. Briefly, 50 µl of 0.1 µg/µl of protein samples was added 96-well plate together with 50 μL of the antibody cocktail. The reaction was incubated at room temperature for 1 hour with shaking at 400 rpm. After incubated, triplicate washing was performed with the 350 μL of 1X washing buffer. Then, TMB solution was added and incubated at room temperature with shaking at 400 rpm for 15 min. At the end of reaction, the stop reaction solution was added, and the optical density was measured by spectrophotometry at 450 nm.
Statistical analysis
The statistical tests were performed using commercially available software (GraphPad Prism). All values were expressed as Mean ± SD. All comparisons were assessed for significance using ANOVA, followed when appropriate by the Tukey-Kramer test. A P-value of less than 0.05 was considered statistically significant.
RESULTS
Simulated ischemia (sI) condition on cardiac cell viability, injury, and intracellular ROS generation
Cardiomyocytes were subjected to simulated ischemia (sI) for various periods of time. The cell viability by MTT assay was performed. The findings demonstrated that a duration of 5 minutes of ischemia resulted in a substantial 20% mortality of cardiomyocytes, compared to the non-ischemic control group (80.40 ± 3.25 % vs 100.0 ± 4.63 %, P< 0.05). Longer duration of ischemia could significantly reduce cell viability (10 minutes 76.81 ± 2.26%, 20 minutes 73.72 ± 3.52%, 30 minutes 64.69 ± 2.70%, 40 minutes 66.64 ± 4.07%, 50 minutes 63.31 ± 3.72%, 60 minutes 52.81 ± 3.26%, 120 minutes 22.86 ± 1.43%) (Figure 1A).
The released-lactate dehydrogenase (LDH) activity was also determined in cardiomyocytes subjected to simulated ischemia (sI) for 5-120 minutes. The results showed that released-LDH activity was significantly increased in sI conditions when compared to control. The longer of the ischemic period, the greater the released-LDH activity (Figure 1B). However, ischemia for 30-120 minutes showed significant increase of released-LDH activity among ischemic group.
Intracellular reactive oxygen species (ROS) was also determined in response to ischemic insult. The results showed that the intracellular ROS level in cardiomyocytes subjected to simulated ischemia (sI) for 5-120 minutes was significantly higher than that of control group. However, ischemia for 30-120 minutes showed significant increase of intracellular ROS level among ischemic group (Figure 1C).
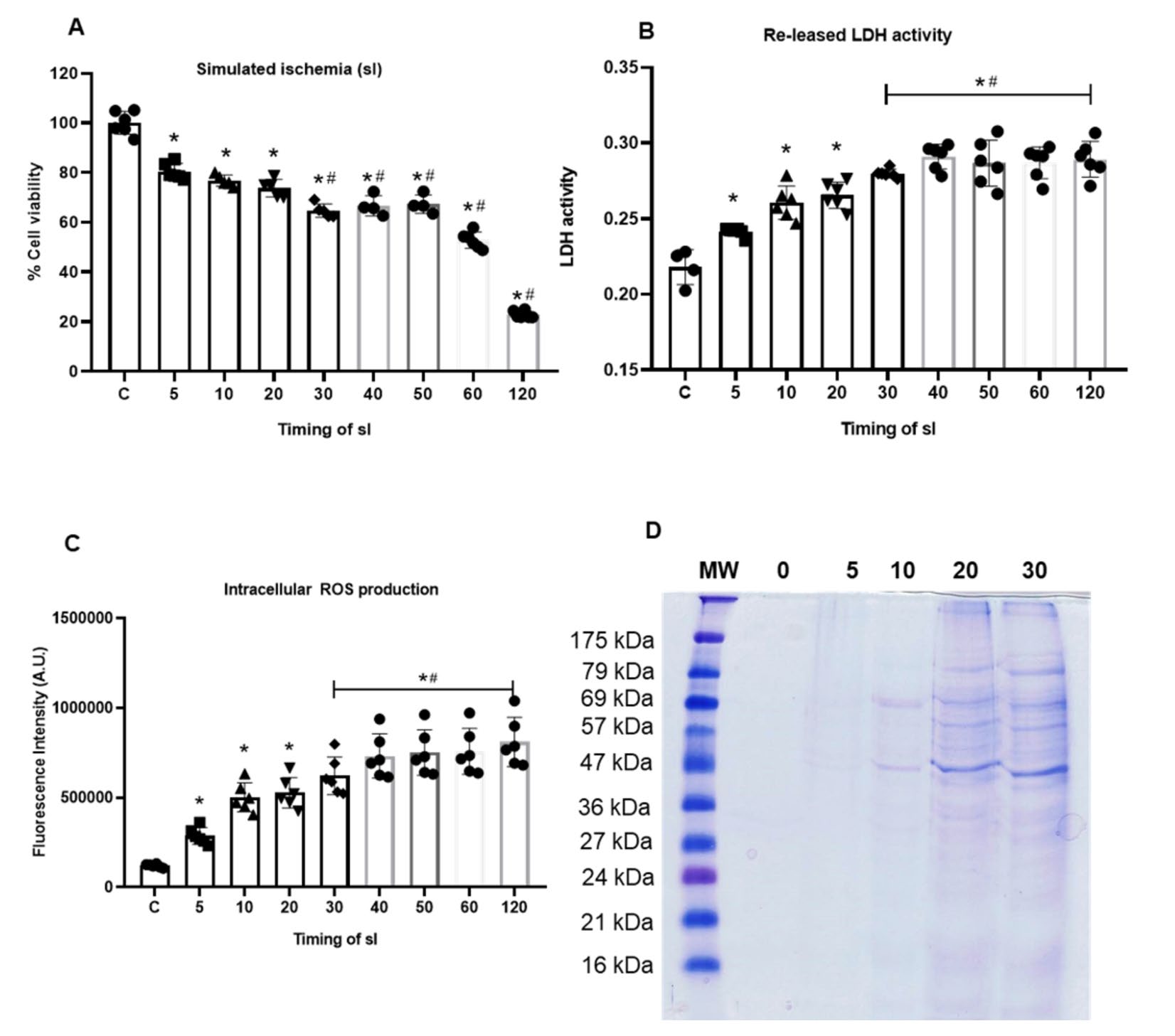
Figure 1. Effect of ischemic duration on cell viability (a), cell injury (b), intracellular ROS generation (c), and proteins in ischemic buffer separated by SDS-PAGE (d).
The simulated ischemic (sI) buffer was collected after cardiomyocytes were subjected to sI for 5, 10, 20, and 30 minutes. The protein in pooled samples were subjected to SDS-PAGE analysis. SDS-PAGE shows predominant bands at molecular weight around 57-69 KDa and 27-36 KDa, which intensity of the band increased corresponding to the ischemic time (Figure 1D).
Identification of cardiac specific protein-DAMPs in response to ischemia
The cardiomyocytes were subjected to 5, 10, 20, and 30 minutes of simulated ischemia (sI). The ischemic buffer each sI duration as well as control sI buffer was collected for proteomic analysis. A total of 14533 differentially expressed proteins (features) in ischemic buffer across the five groups were identified.
To analyze the variation of the individual value from the mean value of the group, the analysis of variance (ANOVA) of the data was performed. The results showed 1561 proteins in ischemic injury were significantly different from control group (Figure 2A).
To directly evaluate the induced protein alterations and determine whether differences in proteomic profiles exist among the groups, pattern recognition techniques including PCA and PLS-DA were both applied to the data set. The results are represented as scores plots, in which a group of samples that displayed a particular proteomic profile, clustered together in a particular area of the plot. The two-dimensional score plots of the PCA and PLS-DA models both demonstrated that the control and 5 minutes of ischemia (I5) were separated from each other and from other ischemic duration (I10, I20, and I30) in the PC1 direction (Figure 2B, C).
Heat maps of the top 30 selected proteins are shown in Figure 2D and table 1. Among those top 30 proteins, analysis by the variable importance in projection (VIP) plot from PLS-DA model, showed ranking the protein based on their importance in discrimination among control and each ischemic duration (Figure 2E).
Lactotransferrin (LTF), also known as Lactoferrin (LF) is considered as the most important protein that is influenced by ischemia. The longer the ischemia, the greater changes could be detected in lactotransferin. The presence of lactoferrin in each duration of ischemia was determined by ELISA. The results showed that the lactoferrin concentrations were significantly increased in ischemic buffer when compared to non-ischemic control (Figure 3).
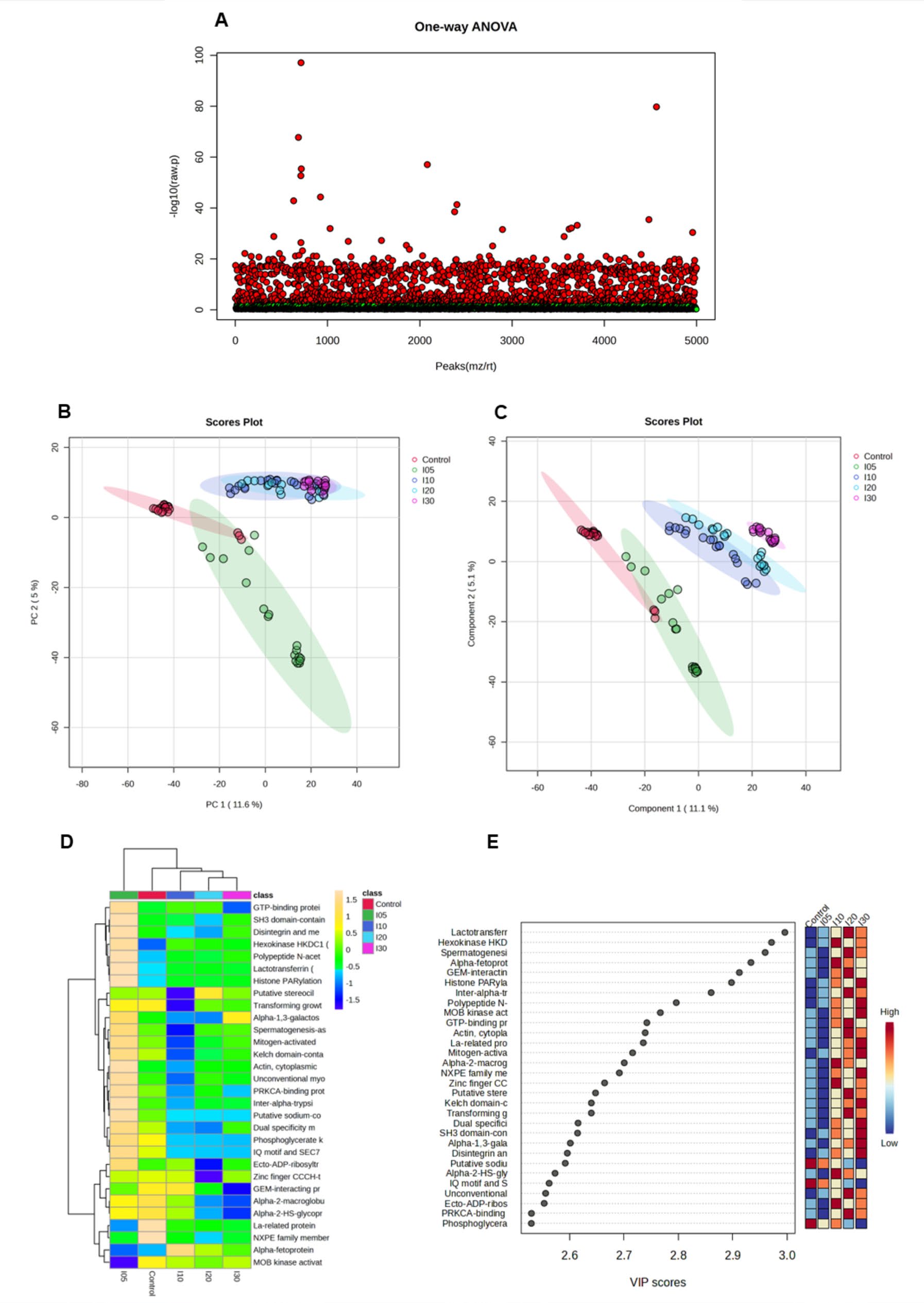
Figure 2. Identification of protein in response to ischemia duration by proteomic analysis. Proteins in ischemic injury vs control group (a), the pattern recognition techniques including PCA (b) and PLS-DA (c), the heat maps of the top 30 selected proteins (d) and the variable importance in projection (VIP) plot from PLS-DA model (e).
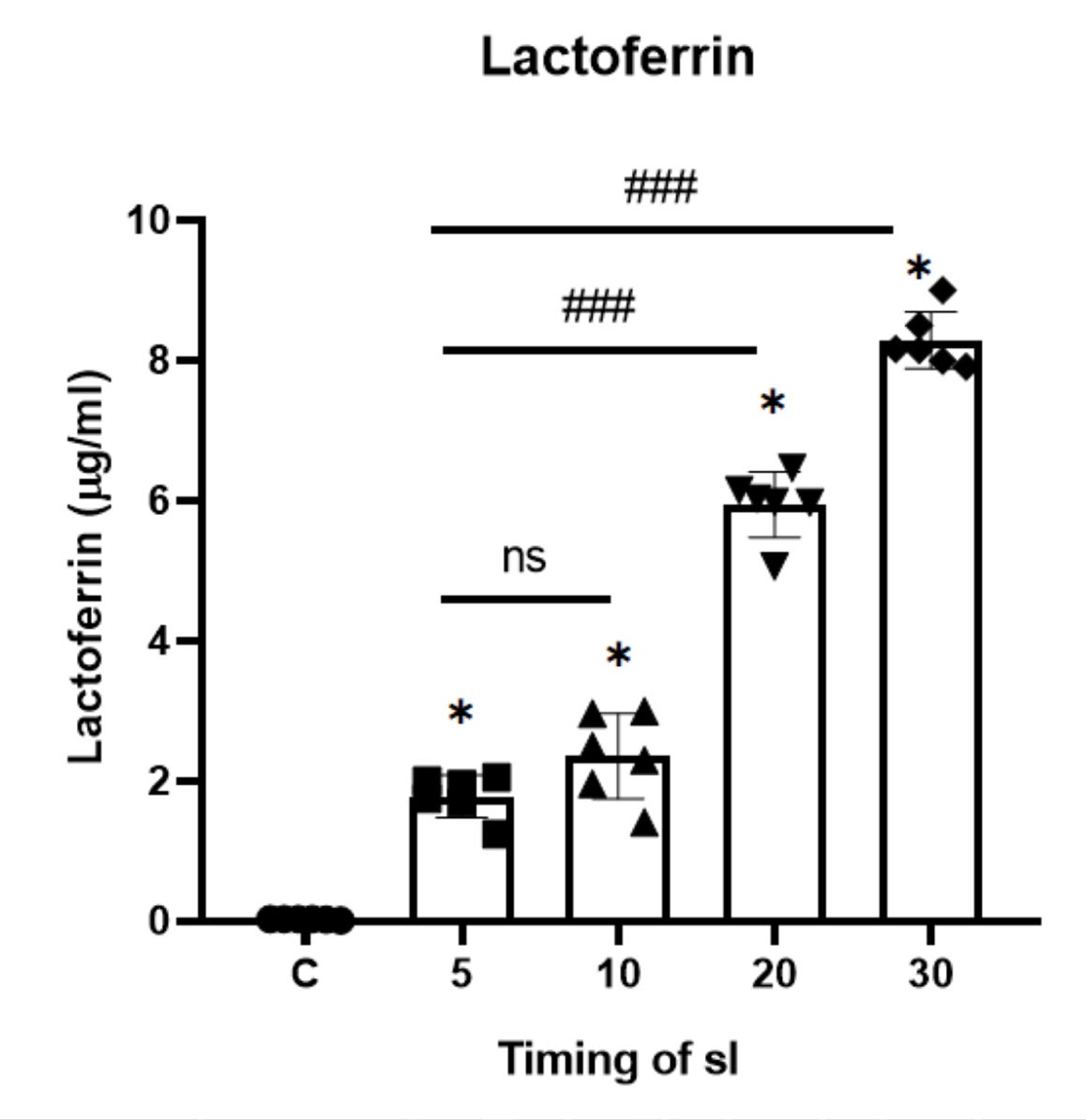
Figure 3. Lactoferrin level in ischemic buffer determined by ELISA.
Pairwise comparisons of the four ischemic durations
To identify differences between each ischemic duration, all four possible pairwise comparisons between each ischemic condition and control (non-ischemia) were carried out in addition to the ANOVA. The expressed proteins with adjusted significance values below the alpha level of 0.05 were considered significant.
The fold-change analysis with volcano plots (Figure 4) indicated that there were 196, 211, 292, 416 proteins statistically significantly higher in ischemic buffer of I5 (Figure 4A, B), I10(Figure 4C, D), I20 (Figure 4E, F) and I30 (Figure 4G, H), when compared to control group, respectively. From the volcano plots, 147 proteins were identified.
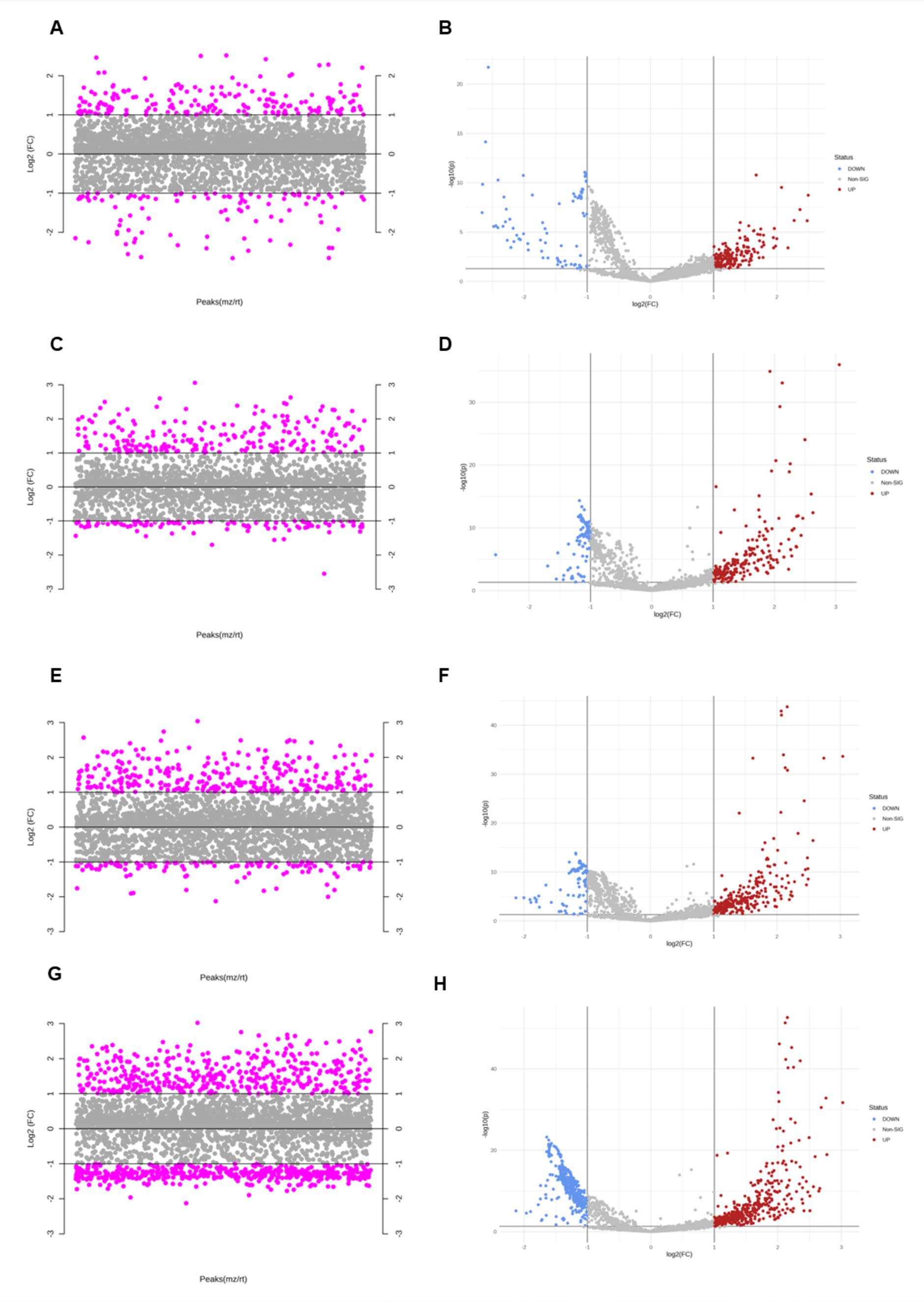
Figure 4. The pairwise comparisons of the four ischemic durations presented in volcano plot in ischemic buffer of I5 (a, b), I10(c, d), I20 (e, f) and I30 (g, h).
Table 1. List to top 30 proteins highly expressed in ischemic injury analyzed by the variable importance in projection (VIP) plot from PLS-DA model.
|
Protein names |
Comp. 1 |
Comp. 2 |
Comp. 3 |
Comp. 4 |
Comp. 5 |
Comp. 6 |
Comp. 7 |
Comp. 8 |
|
Lactotransferrin (Lactoferrin) (EC 3.4.21.-) |
2.9958 |
2.8774 |
2.8023 |
2.7911 |
2.7891 |
2.7887 |
2.7886 |
2.7885 |
|
Hexokinase HKDC1 (EC 2.7.1.1) (Hexokinase domain-containing protein 1) |
2.9713 |
2.8436 |
2.7645 |
2.7534 |
2.7513 |
2.7509 |
2.7508 |
2.7507 |
|
Spermatogenesis-associated protein 2-like protein (SPATA2-like protein) |
2.9596 |
2.8311 |
2.7518 |
2.7409 |
2.7386 |
2.7382 |
2.7381 |
2.738 |
|
Alpha-fetoprotein (Alpha-1-fetoprotein) |
2.9334 |
2.8421 |
2.7669 |
2.7558 |
2.7538 |
2.7534 |
2.7532 |
2.7532 |
|
GEM-interacting protein (GMIP) |
2.9121 |
2.8351 |
2.7562 |
2.7451 |
2.7432 |
2.7428 |
2.7427 |
2.7426 |
|
Histone PARylation factor 1 |
2.8976 |
2.7601 |
2.6857 |
2.6751 |
2.6732 |
2.6728 |
2.6727 |
2.6726 |
|
Inter-alpha-trypsin inhibitor heavy chain H2 (ITI-HC2) |
2.8602 |
2.7177 |
2.6493 |
2.6387 |
2.6365 |
2.6361 |
2.636 |
2.6359 |
|
Polypeptide N-acetylgalactosaminyltransferase 5 (EC 2.4.1.41) |
2.7961 |
2.6971 |
2.6173 |
2.6069 |
2.6051 |
2.6048 |
2.6046 |
2.6046 |
|
MOB kinase activator 3A (MOB-LAK) |
2.7668 |
2.6844 |
2.6078 |
2.6003 |
2.5982 |
2.5979 |
2.5977 |
2.5977 |
|
GTP-binding protein Di-Ras3 |
2.7419 |
2.6452 |
2.5775 |
2.5673 |
2.5657 |
2.5654 |
2.5652 |
2.5652 |
|
Actin, cytoplasmic 2 (EC 3.6.4.-) |
2.7389 |
2.6012 |
2.5403 |
2.5313 |
2.5292 |
2.5288 |
2.5287 |
2.5286 |
|
La-related protein 1B (La ribonucleoprotein domain family member 1B) |
2.7354 |
2.6797 |
2.6139 |
2.6034 |
2.6014 |
2.601 |
2.6009 |
2.6008 |
|
Mitogen-activated protein kinase kinase kinase 9 (EC 2.7.11.25) |
2.7158 |
2.5943 |
2.517 |
2.5079 |
2.507 |
2.5067 |
2.5066 |
2.5065 |
|
Alpha-2-macroglobulin (Alpha-2-M) |
2.7004 |
2.6906 |
2.6344 |
2.624 |
2.6223 |
2.6219 |
2.6218 |
2.6217 |
|
NXPE family member 2 (Protein FAM55B) |
2.6914 |
2.5852 |
2.5385 |
2.5285 |
2.5265 |
2.5261 |
2.526 |
2.5259 |
|
Zinc finger CCCH-type antiviral protein |
2.6643 |
2.5611 |
2.5032 |
2.4944 |
2.4924 |
2.492 |
2.4919 |
2.4918 |
|
Putative stereocilin-like protein (Stereocilin pseudogene 1) |
2.6476 |
2.5537 |
2.4788 |
2.4722 |
2.4712 |
2.4708 |
2.4707 |
2.4707 |
|
Kelch domain-containing protein 8A (Substitute for delta-EGFR expression 1) |
2.6399 |
2.4885 |
2.4377 |
2.428 |
2.427 |
2.4268 |
2.4267 |
2.4266 |
|
Transforming growth factor beta-1-induced transcript 1 protein |
2.6398 |
2.5584 |
2.4832 |
2.4758 |
2.475 |
2.4748 |
2.4747 |
2.4746 |
|
Dual specificity mitogen-activated protein kinase kinase 1 (MAPKK 1) |
2.6154 |
2.5046 |
2.4387 |
2.4294 |
2.4275 |
2.4272 |
2.4271 |
2.4271 |
|
SH3 domain-containing kinase-binding |
2.6145 |
2.4636 |
2.3911 |
2.3822 |
2.3818 |
2.3815 |
2.3814 |
2.3814 |
|
Alpha-1,3-galactosyltransferase 2 |
2.6012 |
2.4594 |
2.3927 |
2.3846 |
2.3829 |
2.3826 |
2.3825 |
2.3824 |
|
Disintegrin and metalloproteinase |
2.5957 |
2.5132 |
2.4401 |
2.4303 |
2.4291 |
2.4289 |
2.4288 |
2.4287 |
|
Putative sodium-coupled neutral amino acid transporter 11 (Solute carrier family 38 member 11) |
2.5918 |
2.4158 |
2.3561 |
2.3480 |
2.3463 |
2.346 |
2.3459 |
2.3459 |
|
Alpha-2-HS-glycoprotein |
2.5731 |
2.6582 |
2.5968 |
2.5864 |
2.5847 |
2.5844 |
2.5842 |
2.5842 |
|
IQ motif and SEC7 domain-containing |
2.5622 |
2.3927 |
2.322 |
2.3130 |
2.311 |
2.3107 |
2.3107 |
2.3106 |
|
Unconventional myosin-VIIb |
2.5558 |
2.4414 |
2.3686 |
2.3591 |
2.3575 |
2.3573 |
2.3572 |
2.3572 |
|
Ecto-ADP-ribosyltransferase 4 |
2.5531 |
2.4359 |
2.3882 |
2.3789 |
2.3778 |
2.3776 |
2.3775 |
2.3774 |
|
PRKCA-binding protein (Protein kinase |
2.5297 |
2.485 |
2.4112 |
2.4038 |
2.4030 |
2.4027 |
2.4025 |
2.4025 |
|
Phosphoglycerate kinase 1 (EC 2.7.2.3) |
2.5295 |
2.3751 |
2.3045 |
2.2954 |
2.2935 |
2.2932 |
2.2931 |
2.2930 |
Identification of candidate proteins for novel early cardiac biomarkers
To identify cardiac specific protein DAMPs as novel early biomarkers, the proteins that were found to highly released from cells to ischemic buffer, as early as 5 minutes of ischemia and sustain until 30 minutes of ischemia. The Venn diagram (Figure 5A) further showed that most of the upregulated proteins associated with ischemic injury from various ischemic durations, which were present in the overlapping portion of the 4 ischemic durations. These results implicated that biologically meaningful of these proteins.
The results showed that there are 16 proteins that were found from 5 minutes of ischemia until 30 minutes of ischemia (table 2). However, those 16 proteins were not identical with the proteins found in the top 30 selected proteins (Figure 5B). Further analysis of proteins that were highly found. From 10-30 minutes of ischemia showed that there are 147 proteins that continuously released from cardiac cells to ischemic buffer from 10-30 minutes of ischemia. Among these sustain released proteins, 21 proteins were found to be identical to the top 30 proteins that impact by ischemia showed in VIP plot from PLS-DA (Figure 5C). Therefore, these 21 proteins are considered as candidates for novel early cardiac biomarkers. Furthermore, the potential interactions of the 21 candidate proteins were analysed using STITCH 5.0. The results showed that, among the 21 candidate proteins, there is interaction found in 15 proteins. The prediction of signaling pathway related regulation of protein serine/threonine kinase activity (GO:0071900, FDR = 7.51e-3) (Figure 5D).
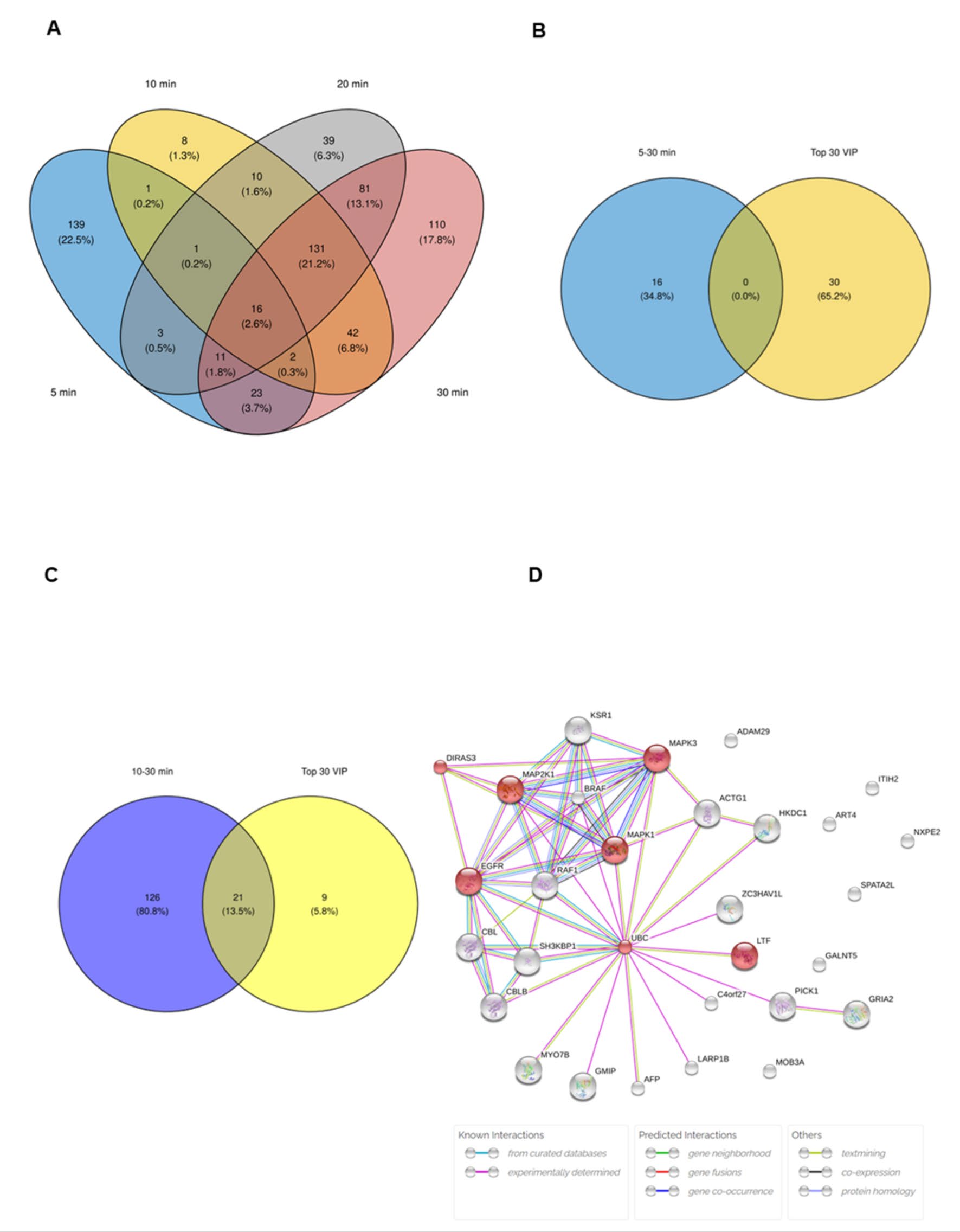
Figure 5. Identification of candidate proteins for novel early cardiac biomarkers. The Venn diagram of most of the upregulated proteins associated with ischemic injury from various ischemic durations (a), the overlapping analysis of proteins that found from 5 -30 minutes of ischemia (b), and 10-30 minutes of ischemia (c). The potential interactions of the 21 candidate proteins were analyzed using STITCH 5.0. (d).
Table 2. List the top 16 proteins that were found from 5 minutes of ischemia to 30 minutes of ischemia.
|
Protein names |
Comp. 1 |
Comp. 2 |
Comp. 3 |
Comp. 4 |
Comp. 5 |
Comp. 6 |
Comp. 7 |
Comp. 8 |
|
Integrin alpha-1 |
2.3843 |
2.2723 |
2.2131 |
2.2044 |
2.2041 |
2.2037 |
2.2036 |
2.2036 |
|
Neuroblast differentiation-associated protein AHNAK (Desmoyokin) |
2.3721 |
2.2100 |
2.1461 |
2.1390 |
2.1372 |
2.1369 |
2.1368 |
2.1367 |
|
Fibrous sheath-interacting protein 2 |
2.3403 |
2.1973 |
2.1445 |
2.1393 |
2.1377 |
2.1375 |
2.1374 |
2.1373 |
|
Ankyrin repeat domain-containing protein 36A |
1.7649 |
1.6475 |
1.6039 |
1.6019 |
1.6024 |
1.6022 |
1.6021 |
1.6021 |
|
Microtubule cross-linking factor 1 |
1.5800 |
1.4845 |
1.514 |
1.5082 |
1.507 |
1.5067 |
1.5067 |
1.5067 |
|
Centromere protein F (CENP-F) |
1.5459 |
1.4414 |
1.4208 |
1.4165 |
1.4154 |
1.4152 |
1.4151 |
1.4151 |
|
Probable ubiquitin carboxyl-terminal hydrolase FAF-Y (EC 3.4.19.12) |
1.4624 |
1.4052 |
1.3943 |
1.3902 |
1.3898 |
1.3897 |
1.3896 |
1.3896 |
|
Midasin (MIDAS-containing protein) |
1.3797 |
1.2865 |
1.2583 |
1.2552 |
1.2544 |
1.2543 |
1.2542 |
1.2542 |
|
Zinc finger CCCH domain-containing protein 4 |
1.2649 |
1.177 |
1.1786 |
1.1929 |
1.1945 |
1.1944 |
1.1944 |
1.1944 |
|
Hemicentin-1 |
1.2493 |
1.1653 |
1.1357 |
1.1314 |
1.1307 |
1.1307 |
1.1306 |
1.1306 |
|
Sterile alpha motif domain-containing protein 9 (SAM domain-containing protein 9) |
1.2390 |
1.1586 |
1.2626 |
1.2671 |
1.2721 |
1.2719 |
1.2718 |
1.2718 |
|
Pericentrin (Pericentrin-B) |
1.1490 |
1.0790 |
1.0592 |
1.0651 |
1.0649 |
1.0647 |
1.0647 |
1.0647 |
|
Uncharacterized protein C4orf54 (Familial obliterative portal venopathy) |
0.9716 |
1.0384 |
1.0101 |
1.0073 |
1.0065 |
1.0064 |
1.0064 |
1.0064 |
|
Neuron navigator 3 (Pore membrane and/or filament-interacting-like protein 1) |
0.7452 |
0.7542 |
0.7639 |
0.7611 |
0.7612 |
0.7611 |
0.7611 |
0.7611 |
|
Zinc finger protein 64 (Zfp-64) |
0.7094 |
0.7104 |
0.7323 |
0.7513 |
0.7510 |
0.7509 |
0.7511 |
0.7510 |
|
Anoctamin-1 |
0.5526 |
0.5273 |
0.5682 |
0.5732 |
0.6008 |
0.6043 |
0.6043 |
0.6043 |
DISCUSSION
In the present study, we identify candidate biomarkers by comprehensively analyzing the proteins released into the condition buffer (simulated ischemic buffer) after various durations of in vitro simulated ischemia (sI) in rat cardiac myoblast cells. The duration of ischemic injury chosen in this study was considered to cause mild cardiac injury, which may represent the early biomarker for ischemic heart disease. In this study, an in vitro simulated ischemia was performed in cardiac cells. The proteomic profiles from this study are, therefore, specific to cardiomyocytes. The proteins that identify to be impacted by ischemic injury and released to circulation could considered as a specific marker of early cardiac injury.
Protein damage-associated molecular patterns (DAMPs) include cardiomyocyte-specific proteins produced after ischemia injury. A quick release and sustained detection are necessary for an optimal cardiac marker because the biomarker must be released into blood rapidly after myocardial damage and persist in circulation to be detected. In this study we found that there were 16 proteins commonly found in ischemic buffer from 5-30 minutes. Although those 16 proteins are not a member of the top 30 proteins that are impacted by ischemia, the information was also found to be interesting and require further validation in an in vivo animal model. In addition, there are 147 proteins released to ischemic buffer from 10 to 30 minutes of ischemia. Among the 147 proteins, 21 proteins were found to be identical to the top 30 proteins that are impacted by ischemia as shown in a VIP plot from PLS-DA. One of these proteins is lactotransferrin or lactoferrin, which is the protein with the highest VIP score and is sustainably found in every ischemic duration. Lactotransferrin (LTF) or lactoferrin is a multifunctional protein present in diverse biological secretions, including milk, tears, saliva, vaginal fluids, and sperm (Wang et al., 2019). It has a broad range of functions, including iron binding/transfer as well as antibacterial, antiviral, antifungal, anti-inflammatory, and anticarcinogenic properties [Wang et al., 2019]. However, the cardio-vascular roles of LTF have only been reported recently. Recent reports indicate that LTF ameliorates pathological cardiac hypertrophy in geriatric rodents (Huang et al., 2021). Moreover, the cardioprotective effect of LTF against myocardial ischemia/reperfusion injury has also been recently reported (Omiya et al., 2022). Although the cardioprotective effect of LTF has been reported, the role of early cardiac biomarkers has never been investigated.
Jacquet et al. (Jacquet et al., 2009) identified cardiac myosin binding protein C (cMyBP-C) as a novel cardiac marker for early ischemia/reperfusion injury in an ex vivo model. Rat hearts underwent 5 minutes of global ischemia followed by reperfused with Krebs-Henseleit buffer. The coronary effluent was collected and analyzed by the proteomic approach based on two-dimensional gel electrophoresis. All protein spots were excised and identified by tandem mass spectrometry. In this study, the researcher had to "hand-select" the proteins of interest, omitting the more prevalent or less fascinating ones. Shotgun proteomics, on the other hand, could provide a greater number of protein identifications (Dowell et al., 2008). Because two-dimensional gel electrophoresis may not identify several proteins in ischemic coronary effluent, few cardiac-specific biomarkers have been found and validated. Jacquet et al. and the present analysis found no shared proteins, possibly due to the distinct study design and proteomic technique.
The global ischemia model may not accurately depict myocardial ischemia in coronary arteries. Atherosclerotic plaque blocking coronary arteries damages the myocardium under the vessel's supply point. Global ischemia model may impact region. The 5-minute ischemia procedure revealed moderate ischemia, however global ischemia might cause greater damage than physiological ischemia. Thus, localized ischemia like LAD coronary artery closure may yield more physiological and therapeutically significant effects. Ex vivo proteome study in lab animals may provide non-cardiomyocyte protein. Cardiomyocytes, cardiac fibroblasts, endothelium, smooth muscle, and immune cells make up the heart. The global ischemia in whole heart experiment procedure may misunderstand proteomic results by recruiting non-cardiomyocyte protein. Selectively produced ischemia damage in cardiomyocytes may filter non-cardiomyocyte markers for more specific information. The current investigation identified cardiomyocyte-only proteins in an in vitro model.
Simulated ischemia (sI) is an in vitro procedure for producing ischemic stress to the cardiac cell and has been used in several studies of in vitro simulated ischemia (Prompunt et al., 2018; Sanit et al., 2019; Mongkolpathumrat et al., 2022). This stress could be comparable with real ischemia/reperfusion injury that occur in ex vivo or in vivo models. The model of simulated ischemia used in this experiment was based on the metabolic blockade also called “chemical ischemia.” The ischemic buffer contains 2-deoxyglucose, Na lactate, and Na dithionite. The 2-deoxyglucose is a glucose molecule, which 2-hydroxy group replaced by hydrogen molecule. Therefore, it could not undergo further glycolysis, mimicking the ischemic condition (Diaz et al., 2014). During ischemia, cells undergo cellular acidosis because the H+ increases. Therefore, Na lactate, which was added in ischemic buffer, induced the cell to acidosis condition (Diaz et al., 2014). Na dithionite is an oxygen scavenger which captures oxygen molecules in the solution (Juven and Rosenthal, 1985; Tao et al., 2008). The cell in this experiment was treated with an ischemic buffer and also lacked oxygen and glucose. This was similar to the real ischemia that occurs in humans. The various ischemic duration was induced by incubation of cardiac cells with an ischemic buffer. In this study, incubation with ischemic buffer for 5 to 10 minutes could be considered to be early phase or mild injury, which is sufficient to increase ROS generation and cause approximately 20% cardiac cell death (Figure 1A-C). The global ischemia for 5 minutes, in an ex vivo model of Langendorff perfusing heart, caused around 5% of infarct size. The longer the ischemic period, the greater the infarct area (Jacquet et al., 2009).
Our study has various limitations that should be taken into account. The present work offers empirical evidence supporting the idea that LTF might serve as a potential biomarker for detecting early ischemia damage, especially in human cardiomyocytes. While previous studies have demonstrated the therapeutic benefits of LTF in ischemia/reperfusion injury in in vivo animal model, our research reveals a novel function of LTF as a potential early indicator during the ischemic phase of injury. Hence, the present result is regarded as evidence that necessitates more investigation and validation in an in vivo model and clinical trial, respectively. Utilising animal cells and doing additional research in an ex vivo or in vivo experimental animal model, similar to earlier studies, appears to be a valid sequential study paradigm. However, conducting the experiment utilising human cell sources is challenging for further investigation. While the AC16 cells possess biological characteristics that resemble those of human cardiomyocytes, they do not exhibit electrical qualities. Experimentally, isolating human cardiomyocytes is deemed unfeasible. Thus, utilising more pertinent models, such as those in the undamaged rat or mouse heart, might yield more accurate functional data, as these models closely resemble the actual physiological processes in the heart. However, it is crucial to exercise caution and meticulous analysis when conducting clinical trials using these models. Moreover, it is crucial to assess the clinical use of new identified biomarkers, considering their analytical sensitivity, specificity, and potential in comparison to other established biomarkers, before advancing to clinical trials.
CONCLUSION
In conclusion, the present study identified lactotransferrin (LTF) as an early protein damage associated molecular pattern (DAMPs) released specifically from cardiac cell subjected to ischemic injury. Further studies should be focused on in vivo validation of potential protein markers that could be used in practical clinical applications.
ACKNOWLEDGEMENTS
We would like to thank for research facilities and equipment were supported by the Integrative Biomedical Research Unit (IBRU), Faculty of Allied Health Sciences, Naresuan University, Biomedical Engineering Institute, Chiang Mai University, and National Centre for Genetic Engineering and Biotechnology (BIOTEC), National Science and Technology Development Agency Thailand.
AUTHOR CONTRIBUTIONS
Sarawut Kumphune: Writing – review & editing, Writing – original draft, Validation, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization. Nitchawat Paiyabhrom: Writing – original draft, Validation, Methodology, Formal analysis, Data curation. Porrnthanate Seenak: Validation, Methodology, Formal analysis, Data curation. Worawat Songjang: Validation, Methodology, Formal analysis, Data curation. Arunya Jiraviriyakul: Methodology, Investigation, Funding acquisition, Data curation, Conceptualization. Noppadon Jumroon: Writing – original draft, Formal analysis. Panyupa Pankhong: Methodology, Formal analysis, Data curation. Siriwan Thaisakun: Validation, Formal analysis, Data curation. Narumon Phaonakrop: Validation, Formal analysis, Data curation. Sittiruk Roytrakul: Validation, Methodology, Formal analysis, Data curation. Nitirut Nernpermpisooth: Writing – review & editing, Writing – original draft, Supervision, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization.
CONFLICT OF INTEREST
The authors declare that they hold no competing interests.
REFERENCES
Bardou, P., Mariette, J., Escudie, F., Djemiel, C., and Klopp, C. 2014. Jvenn: An interactive Venn diagram viewer. BMC Bioinformatics. 15: 293.
Charpentier, S., Ducasse, J. L., Cournot, M., Maupas-Schwalm, F., Elbaz, M., Baixas, C., Juchet, H., Lang, T., and Lauque, D. 2010. Clinical assessment of ischemia-modified albumin and heart fatty acid-binding protein in the early diagnosis of non-ST-elevation acute coronary syndrome in the emergency department. Academic Emergency Medicine. 17: 27-35.
Diaz, R. J., Harvey, K., Boloorchi, A., Hossain, T., Hinek, A., Backx, P. H., and Wilson, G. J. 2014. Enhanced cell volume regulation: A key mechanism in local and remote ischemic preconditioning. American Journal of Physiology-Cell Physiology. 306: C1191-1199.
Dowell, J. A., Frost, D. C., Zhang, J., and Li, L. 2008. Comparison of two-dimensional fractionation techniques for shotgun proteomics. Analytical Chemistry. 80: 6715-6723.
Howe, E. A., Sinha, R., Schlauch, D., and Quackenbush, J. 2011. RNA-Seq analysis in MeV. Bioinformatics. 27: 3209-3210.
Huang, L., Chen, R., Liu, L., Zhou, Y., and Chen, Z. 2021. Lactoferrin ameliorates pathological cardiac hypertrophy related to mitochondrial quality control in aged mice. Food & Function. 12: 7514-7526.
Jacquet, S., Yin, X., Sicard, P., Clark, J., Kanaganayagam, G. S., Mayr, M., and Marber, M. S. 2009. Identification of cardiac myosin-binding protein C as a candidate biomarker of myocardial infarction by proteomics analysis. Molecular & Cellular Proteomics. 8: 2687-2699.
Jennings, R. B. and Reimer, K. A. 1991. The cell biology of acute myocardial ischemia. Annual Review of Medicine. 42: 225-246.
Johanson, P., Wagner, G. S., Dellborg, M., and Krucoff, M. W. 2003. ST-segment monitoring in patients with acute coronary syndromes. Current Cardiology Reports. 5: 278-283.
Juven, B. J., and Rosenthal, I. 1985. Effect of free-radical and oxygen scavengers on photochemically generated oxygen toxicity and on the aerotolerance of Campylobacter jejuni. The Journal of Applied Bacteriology. 59: 413-419.
Land, W. G. 2015. The role of damage-associated molecular patterns (DAMPs) in human diseases: Part II: DAMPs as diagnostics, prognostics and therapeutics in clinical medicine. Sultan Qaboos University Medical Journal. 15: e157-170.
Lowry, O. H., Rosebrough, N. J., Farr, A. L., and Randall, R. J. 1951. Protein measurement with the Folin phenol reagent. Journal of Biological Chemistry. 193: 265-275.
Mi, H., Muruganujan, A., Ebert, D., Huang, X., and Thomas, P. D. 2019. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Research. 47: D419-D426.
Mongkolpathumrat, P., Kijtawornrat, A., Suwan, E., Unajak, S., Panya, A., Pusadee, T., and Kumphune, S. 2022. Anti-protease activity deficient secretory leukocyte protease inhibitor (SLPI) exerts cardioprotective effect against myocardial ischaemia/reperfusion. Biomedicines. 10(5): 988.
Mozaffarian, D., Benjamin, E. J., Go, A. S., Arnett, D. K., Blaha, M. J., Cushman, M., Das, S. R., de Ferranti, S., Despres, J. P., Fullerton, H. J., et al. 2016. Heart disease and stroke statistics-2016 update: A report from the american heart association. Circulation. 133: e38-360.
Nernpermpisooth, N., Prompunt, E., and Kumphune, S. 2017. An in vitro endothelial cell protective effect of secretory leukocyte protease inhibitor against simulated ischaemia/reperfusion injury. Experimental and Therapeutic Medicine. 14: 5793-5800.
Omiya, K., Nakadate, Y., Oguchi, T., Sato, T., Matsuoka, T., Abe, M., Kawakami, A., Matsukawa, T., and Sato, H. 2022. Cardioprotective effects of enteral vs. parenteral lactoferrin administration on myocardial ischemia-reperfusion injury in a rat model of stunned myocardium. BMC Pharmacology and Toxicology. 23: 78.
Paiyabhroma, N., Nernpermpisooth N., Kumphune, S. 2018. The recombinant human secretory leukocyte protease inhibitor (SLPI) protects cardiac fibroblasts injury against an in vitro ischemia/reperfusion injury. Journal of Applied Pharmaceutical Science. 8: 7.
Pang, Z., Zhou, G., Ewald, J., Chang, L., Hacariz, O., Basu, N., and Xia, J. 2022. Using MetaboAnalyst 5.0 for LC-HRMS spectra processing, multi-omics integration and covariate adjustment of global metabolomics data. Nature Protocols. 17: 1735-1761.
Prompunt, E., Nernpermpisooth, N., Sanit, J., and Kumphune, S. 2018. Overexpression and pre-treatment of recombinant human secretory leukocyte protease inhibitor (rhSLPI) reduces an in vitro ischemia/reperfusion injury in rat cardiac myoblast (H9c2) cell. Biomolecular Concepts. 9: 17-32.
Roh, J. S. and Sohn, D. H. 2018. Damage-associated molecular patterns in inflammatory diseases. Immune Network. 18: e27.
Sanit, J., Prompunt, E., Adulyaritthikul, P., Nokkaew, N., Mongkolpathumrat, P., Kongpol, K., Kijtawornrat, A., Petchdee, S., Barrere-Lemaire, S., and Kumphune, S. 2019. Combination of metformin and p38 MAPK inhibitor, SB203580, reduced myocardial ischemia/reperfusion injury in non-obese type 2 diabetic Goto-Kakizaki rats. Experimental and Therapeutic Medicine. 18: 1701-1714.
Szklarczyk, D., Santos, A., von Mering, C., Jensen, L. J., Bork, P., and Kuhn, M. 2016. STITCH 5: Augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Research. 44: D380-D384.
Tao, Z., Goodisman, J., and Souid, A. K. 2008. Oxygen measurement via phosphorescence: reaction of sodium dithionite with dissolved oxygen. The Journal of Physical Chemistry A. 112: 1511-1518.
Tyanova, S., Temu, T., and Cox, J. 2016. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nature Protocols. 11: 2301-2319.
Tyanova, S., Temu, T., Sinitcyn, P., Carlson, A., Hein, M. Y., Geiger, T., Mann, M., and Cox, J. 2016. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nature Methods. 13: 731-740.
Wang, B., Timilsena, Y. P., Blanch, E., and Adhikari, B. 2019. Lactoferrin: Structure, function, denaturation and digestion. Critical Reviews in Food Science and Nutrition. 59: 580-596.
Yellon, D. M. and Hausenloy, D. J. 2007. Myocardial reperfusion injury. The New England Journal of Medicine. 357: 1121-1135.
Zhao, T. and Wang, Z. 2022. GraphBio: A shiny web app to easily perform popular visualization analysis for omics data. Frontiers in Genetics. 13: 957317.
OPEN access freely available online
Natural and Life Sciences Communications
Chiang Mai University, Thailand. https://cmuj.cmu.ac.th
Sarawut Kumphune 1,2,3, Nitchawat Paiyabhroma3,4, Porrnthanate Seenak 3,5, Worawat Songjang 3,4, Arunya Jiraviriyakul 3,4, Noppadon Jumroon 3,4, Panyupa Pankhong 3,4, Siriwan Thaisakun 6, Narumon Phaonakrop 6, Sittiruk Roytrakul 6, and Nitirut Nernpermpisooth 3,5,*
1 Biomedical Engineering and Innovation Research Centre, Chiang Mai University, Chiang Mai, 50200 Thailand.
2 Biomedical Engineering Institute, Chiang Mai University, Chiang Mai, 50200 Thailand.
3 Integrative Biomedical Research Unit (IBRU), Faculty of Allied Health Sciences, Naresuan University, Phitsanulok, 65000 Thailand.
4 Department of Medical Technology, Faculty of Allied Health Sciences, Naresuan University, Phitsanulok, 65000 Thailand.
5 Department of Cardio-Thoracic Technology, Faculty of Allied Health Sciences, Naresuan University, Phitsanulok, 65000 Thailand.
6 National Centre for Genetic Engineering and Biotechnology (BIOTEC), National Science and Technology Development Agency, Pathum Thani, 12120 Thailand.
Corresponding author: Nitirut Nernpermpisooth, E-mail: nitirutn@nu.ac.th
Total Article Views
Editor: Veerasak Punyapornwithaya,
Chiang Mai University, Thailand
Article history:
Received: May 9, 2024;
Revised: June 24, 2024;
Accepted: June 27, 2024;
Online First: July 3, 2024

