
Improved PCR for Detection of Xanthomonas euvesicatoria pv. perforans in tomato seeds
Jutatape Watcharachaiyakup*, Kanchanaphon Sawangchaitham, Parichart Burns3 and Wichai Kositratana*Published Date : July 10, 2023
DOI : https://doi.org/10.12982/NLSC.2023.054
Journal Issues : Number 3, July-September 2023
Abstract The detection efficiency of the PCR methods for Xanthomonas euvesicatoria pv. perforans (Xep) was determined. Xep specific primer set; HpaF-f/HpaF-r was compared with the previously reported Bs-XpF/Bs-XpR primers in multiplex PCR reaction and singleplex PCR for detection of Xep were tested. The improvement of PCR amplification efficacy was investigated by adding PCR additives or either 5% DMSO or 1 M betaine. Adding both PCR additives gave the same lowest detection limit of Xep detection in both multiplex and singleplex PCR at 100 fg. However, multiplex PCR gave no equal amplification rate for both sets of primers and also showed cross-amplification to X. vesicatoria. Then singleplex PCR with both PCR additives was further tested in a tomato seed lot. The artificially inoculated tomato seeds with various concentrations of Xep from 1.05 x 105 to 101 CFU/ml were tested with singleplex PCR. The lowest detection limit was 101 CFU/ml of the 5 replications of each inoculated seed. One artificially inoculated tomato seed from each bacterial cell suspension was mixed with 2,000 healthy seeds making 0.05% (1/2,000) contaminated tomato seeds sample and further tested with singleplex PCR, results showed the detection limit of 101 CFU/ml in 0.05% Xep contaminated seed lot.
Keywords: PCR additives, Seed, Bacterial leaf spot, Tomato, Conventional PCR
Citation: Watcharachaiyakup, J., Sawangchaitham, K., Burns, P., and Kositratana, W. 2023. Improved PCR for Detection of Xanthomonas euvesicatoria pv. perforans in tomato seeds. Natural and Life Sciences Communications. 22(3): e2023054.
INTRODUCTION
Bacterial leaf spot (BLS) is an important disease in members of the Solanaceae family, including pepper (Capsicum annuum) and tomato (Lycopersicon esculentum). The symptoms of the disease in infected leaves, stems and fruits are necrotic lesions and defoliation (Utami et al., 2022). Infected plants with severe symptoms display a leaf blight appearance. BLS disease causes severe yield reduction and damage to tomato and pepper fruits. The agents causing BLS are Xanthomonas spp. including X. euvesicatoria pv. euvesicatoria (Xee), X. euvesicatoria pv. perforans (Xep), X. hortorum pv. gardneri (Xhg) and X. vesicatoria (Xv) (Constantin et al., 2016; Morinière et al., 2020). Due to the severity of the BLS disease and the means of transmission, BLS pathogens including X. euvesicatoria pv. euvesicatoria (Xee) and X. euvesicatoria pv. perforans (Xep) were placed on the European and Mediterranean Plant Protection Organization (EPPO) A2 list of quarantine pests (https://www.eppo.int/ACTIVITIES/plant_quarantine/A2_list).
BLS pathogenic bacteria are introduced to plant aerial surfaces through water and wind (Ryan et al., 2011). The bacteria penetrated the plant hosts through natural openings or wounds. After multiplication to a sufficient population, the bacteria move to the mesophyll tissues and cause symptoms of the disease. In addition, the bacteria can also move to seeds, causing seed-borne transmission (Dutta et al., 2014). Xanthomonas spp. associated with BLS disease are widely distributed in many countries, including Australia, the USA, Spain, India, Iran, Thailand and Australia (Osdaghi et al., 2017; Roach et al., 2018; Sitthitanasin et al., 2020). In Thailand, X. campestris pv. vesicatoria was reported in pepper and tomato (Sriwilai, 1994). It was later reclassified as Xep and Xee based on physiological, biochemical and multilocus sequence analysis of the gene gyrB, efp, dnaK, and atpD (Sitthitanasin et al., 2020). The detection methods for these pathogens were mostly based on conventional polymerase chain reaction (PCR) and real-time PCR using specific primers (Araujo et al., 2012; Jones et al., 1993; Koenraadt et al., 2009; Leite Jr et al., 1995; Lue et al., 2010; Ning, 2012; Pečenka et al., 2020). Four sets of amplified fragment length polymorphism (AFLP) primers from Bazilian isolate including Bs-XeF/Bs-XeR, Bs-XvF/Bs-XvR, Bs-XgF/Bs-XgR, and Bs-XpF/Bs-XpR, were used to identify Xee, Xv, Xhg and Xep, respectively (Koenraadt et al., 2009; Osdaghi et al., 2017; Roach et al., 2018; Scortichini et al., 2013). However, the Bs-XpF / Bs-XpR primer set was unable to amplify the Xep strains of Thailand and Australia (Osdaghi et al., 2017; Sitthitanasin et al., 2020). These led to the development of an effective method for Xep strain in Thailand.
Several factors affect PCR efficiency, including DNA/RNA purity, primer specificity, PCR components, and the presence of contaminants (Sambrook & Russell, 2001). The latter can the potential to be PCR inhibitors. Compositions of plant materials reportedly impacted nucleic acid recovery, purity and contaminants (Hills & Van Staden, 2002; Japelaghi et al., 2011). Phenolic compounds or polysaccharides from the grapevine petiole extracts are PCR inhibitors that interfere with PCR amplification unless the samples were diluted 100-fold (Minsavage et al., 1994). PCR inhibitors: tannins and phenolic compounds are also found in seeds (Walcott, 2003). When used for detection, pathogens can easily be missed. To enhance the efficiency of PCR additives such as dimethyl sulfoxide (DMSO), glycerol, formamide, bovine serum albumin (BSA), nonionic detergents, ammonium sulfate, N,N,N-trimethylglycine (betaine) were added to the PCR mixture (Elizabeth Pelt-Verkuil, 2008). PCR amplification of ITS2 DNA barcodes from 12 species of plants from 12 different families which previously showed no amplifications under standard PCR conditions were enhanced by 91.6% and 75% by adding DMSO and betaine respectively (Varadharajan et al., 2021). Betaine at 1-2 mol/L improved PCR amplification in multiple GC-rich rice DNA segments and multiple DNA polymerases (Wang et al., 2018). Colony PCR for the screening of bacteria using degenerate primer was achieved by adding 3% DMSO and 1 M betaine (Sheu et al., 2000). Acidic polysaccharides: dextran sulfate which is a PCR inhibitor found in spinach at the ratio 50:1 of DNA, PCR amplification could be achieved by adding 0.25% or 0.5% Tween 20, 5% DMSO, or 5% polyethylene glycol 400 (Demeke and Adams, 1992).
BLS disease is a seed-borne transmission. Therefore, a sensitive and accurate method is essential for detecting BLS pathogens in plant health services. Improve Xep detection in tomato seed lots using two PCR additive supplements (DMSO and betaine) was investigated in this study. The detection limit was determined using a serial dilution of template DNA from Xep and artificially inoculated seeds in tomato seed lots with various concentrations of bacterial cells. The detection specificity was tested with various Xep strains and Xanthomonas spp.
MATERIALS AND METHODS
Plant and microbial materials
Tomato (Lycopersicon esculentum) cv. Sidatip-4 seeds obtained from the Thailand Vegetable Research Center (TVRC) were used in this experiment. Genomic DNA from BLS causing bacteria including Xee isolate NCPPB2968T, NCPPB2573, Xv isolate NCPPB422T, NCPPB1332, NCPPB1421, Xhg isolate NCPPB881T, NCPPB4323 and Xep isolate NCPPB4322, NCPPB4321T were purchased from National Collection of Plant Pathogenic Bacteria (NCPPB) (https://www.fera.co.uk/ncppb-ordering-strains). The Xep DOA-1642 culture was provided by the Department of Agriculture, Ministry of Agriculture and Cooperatives, Thailand.
Sensitivity and specificity to Xep strain
The PCR components included 1X AccustartTM II PCR ToughMix® (Quantabio, USA), 0.2 µM forward and reverse primers (Table 1) and 1 µl of DNA template in a total reaction of 10 µl. The PCR profile was 1 cycle of 94 °C for 3 min, 35 cycles of 94 °C for 30 sec, 68°C for 15 sec and 72°C for 40 sec and 1 cycle of 72°C for 60 sec. The presence of bacteria in the samples was determined using bacterial 16S rRNA universal primers fD2/rP1 (Weisburg et al., 1991). Singleplex PCR for Xep detection, HpaF-f/HpaF-r primer set was employed (Ning, 2012). The reference strain NCPPB4332 DNA prepared in 10-fold serial dilution (1 ng to 1 fg) was used as a DNA template. Multiplex PCR components included 1X AccustartTM II PCR ToughMix® (Quantabio, USA), 0.2 µM primers of HpaF-f/HpaF-r (Ning, 2012) and Bs-xgF/Bs-xgR, Xhg specific primers (Koenraadt et al., 2009), 1 µl of DNA template in the total reaction of 10 µl. PCR profile was 1 cycle of 94°C for 3 min, 35 cycles of 94°C for 30 sec, 60°C for 15 sec and 72°C for 40 sec and 1 cycle of 72°C for 60 sec. The Xep reference strain Xep NCPPB4332 and the reference strain Xhg NCPBB881 DNA prepared in 10-fold serial dilution (1 ng to 1 fg) were used as the DNA template. The effectiveness of two additives (5% DMSO and 1M betaine) either alone or combined in enhancing sensitivity and specificity of Xep detection was determined in singleplex PCR and multiplex PCR.
The specificity of singleplex and multiplex PCR specificity for Xep strain was determined using DNA templates from Xee (NCPPB2968T, NCPPB2573), Xv (NCPPB422T, NCPPB1332, NCPBB1421), Xhg (NCPBB881T, NCPBB4323), Xep (NCPBB4322, NCPPB4321T) and related species: X. oryzae pv oryzae (Xoo), X. oryzae pv. oryzicola (Xoc), and X. citri (Xcc). An equal ratio DNA mixture of Xee, Xep, Xv and Xhg was used as a positive control. PCR products were electrophoresed on 1% TBE agarose gel at 100 V for 30 min. The agarose gels were stained in 1 µg/ml of ethidium bromide solution and observed under a UV illuminator.
Detection of the Xep strain in artificially inoculated tomato seeds and contaminated tomato seed lots
Xep DOA-1642 was grown on nutrient agar (beef extract 3 g/L, peptone 5 g/L and agar 15 g/L) for 48 hours at 30°C. The bacterial colony was scraped and used to prepare bacterial suspension in potassium phosphate buffer pH 7.4 (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4). The optical density was measured at a wavelength of 600 nm using a spectrophotometer (Spectronic 20+, Milton Roy, USA) and adjusted to 0.2. The bacterial suspension was diluted 10-fold from 10-3 to 10-6 (101 to 104 CFU/ml). Thirty milliliters of each concentration of bacterial cell suspension were mixed with 1 g of tomato seeds and vacuumed for 60 min with 15 min intervals on and 15 min off. Subsequently, the tomato seeds were soaked in the bacterial cell suspension for a period of 14-16 h and then left to dry naturally at room temperature. Artificially inoculated seeds were kept at room temperature and used within 1 month (modified from Hadas et al. (2005)). The contaminated seed lot was done by adding one artificially inoculated tomato seed from each concentration of bacteria into 2,000 tomato seeds which made 0.05% contaminated seed.
Five inoculated tomato seeds of each bacterial concentration were randomly selected from each bacteria concentration. Each seed was soaked in 1 ml of XCV medium (peptone 10 g/L, potassium bromide 10 g/L, boric acid 0.10 g / L, anhydrous calcium chloride 0.25 g/L, 0.1% tween 80, benomyl 1.5 mg/ml) and shaken at 150 rpm for 1 h at room temperature. The inoculated seed was removed, and the seed-soaked solution was further shaken for 20-24 h. The bacterial pellet was precipitated by centrifuging the seed-soaked solution at 12,396 g (Eppendorf 5418, Eppendorf, USA) for 20 min. DNA was extracted from the bacterial pellet using the PrestoTM Mini gDNA Bacteria Kit (Geneaid, Taiwan), following the manufacturer’s recommendation.
Bacteria from each contaminated seed lot was enriched by soaking 5 lots of seeds (5 x 2,000 seeds) (ISTA, 2019) in 30 ml of XCV media and shaking at 150 rpm at room temperature for 1 hr. After removing the seeds, the seed-soaked solution was shaken for 20-24 hours and centrifuged at 12,987 g (Kubota 7930, Japan) for 20 min. The pellet was used for DNA extraction as mentioned above. The experiments were carried out in triplicate.
Singleplex PCR was carried out using the universal bacteria16S primer and the Xep specific primer. PCR products were electrophoresed on 1% TBE agarose gel at 100 V for 30 min. The agarose gels were stained in 1 µg/ml of ethidium bromide solution and observed under a UV illuminator.
Table 1. List of primers used in this study.
|
Name |
Sequences |
Specificity |
Expected size (bp) |
Sources |
|
HpaF-f |
5’-GTGGCAGGCAGGCAATCGACG-3’ |
Xep |
300 |
Ning, 2012 |
|
HpaF-r |
5’-CCGGCACGTCGACGCCTGGAAACC-3’ |
|
||
|
Bs-xgF |
5’-TCAGTGCTTAGTTCCTCATTGTC-3’ |
Xhg |
154 |
Koenraadt |
|
Bs-xgR |
5’-TGACCGATAAAGACTGCGAAAG-3’ |
|
||
|
fD2 |
5’-CCGAATTCGTCGACAACAGAGTTTG |
16S rRNA |
1,500 |
Weisburg |
|
rP1 |
5’-CCCGGGATCCAAGCTTACGGCTACCTT |
|
|
|
RESULTS
PCR optimization, sensitivity and specificity
Multiplex PCR and singleplex PCR were determined for the detection of Xep and Xhg in tomato seeds. Multiplex PCR could detect Xep at 1 fg and Xhg at 1 pg. The addition of both PCR additives had effects on sensitivity giving the highest sensitivity at 100 fg. The addition of additives, either 1M betaine or 5% DMSO gave a detection level of 1 pg. (Figure 1A). Singleplex PCR with 1 M betaine and 5% DMSO showed the lowest detection limit at 100 fg. Similar results were observed with the addition of 5% DMSO. By adding only 1 M betaine or without any additive the PCR was 10 times less sensitive at 1 pg (Figure 1B).
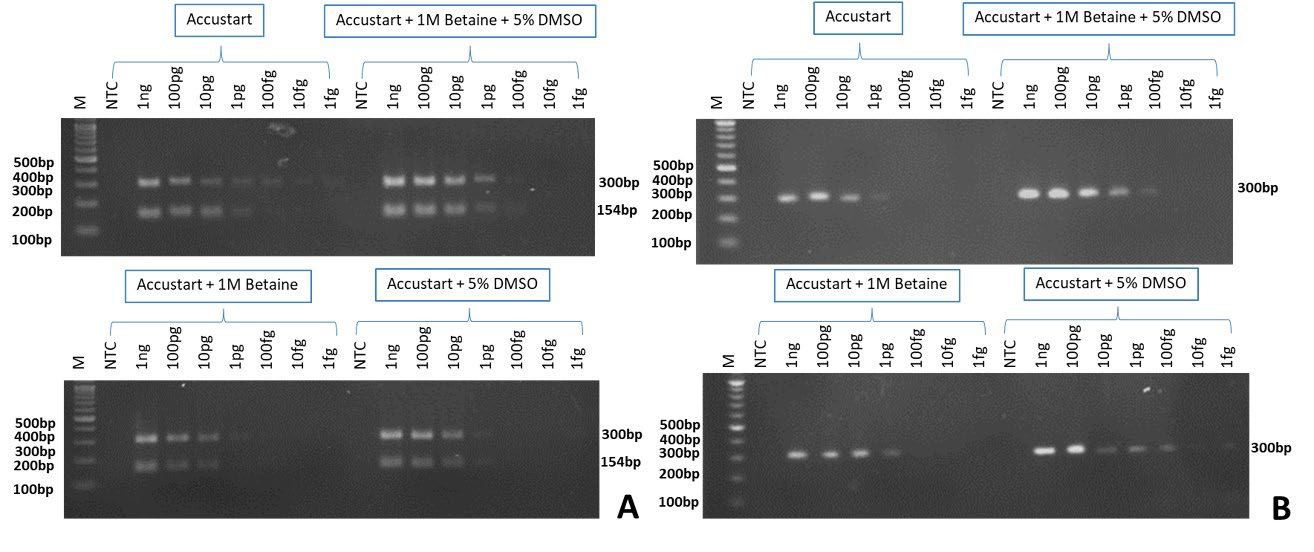
Figure 1. Multiplex (A) and singleplex-PCR (B) of Xanthomonas euvesicatoria pv. perforans; Xep (300 bp) and X. hortorum pv. gardneri; Xhg (154 bp). Ten-fold serial dilution of Xep and Xhg DNA was used as DNA templates from 1 ng to 1 fg. Reactions consisted of those without any additives (Accustart), with 1 M betaine or 5% DMSO and with both additives. M= 100 bp DNA marker (ACTGeneTM, USA), NTC= non-template control.
The specificity of the Xep and Xhg primers was determined by multiplex and singleplex PCR with the addition of two additives (1 M betaine and 5% DMSO). Multiplex PCR produced PCR products of 300 base pairs from template DNA of Xep, Xee, Xv and other Xanthomonas spp. including Xoo, Xoc and Xcc (Figure 2A). The singleplex PCR showed amplification of only the target DNA of Xep (Figure 2B).
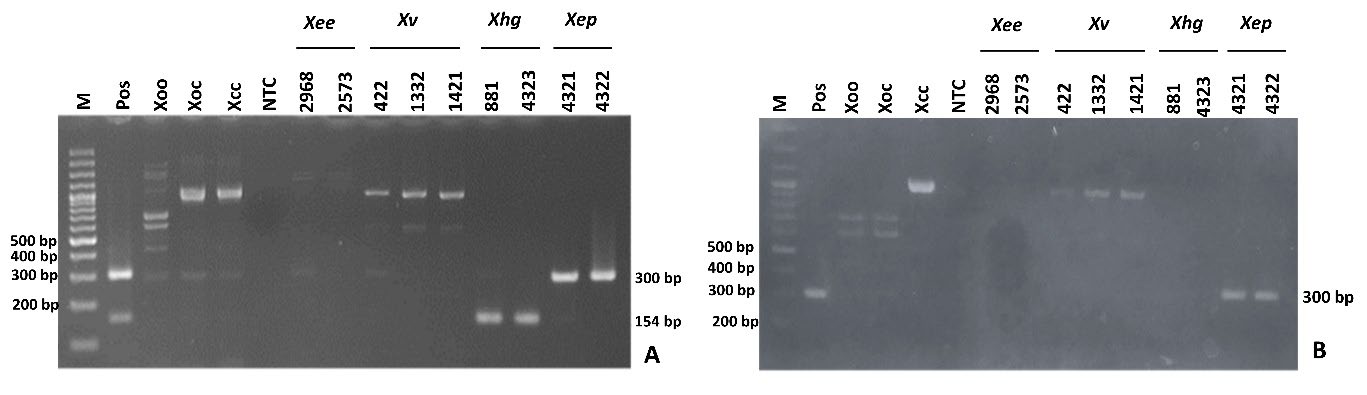
Figure 2. Multiplex (A) and singleplex-PCR (B) of Xanthomonas euvesicatoria pv. perforans (300 bp) and X. hortorum pv. gardneri (154 bp). Reference DNA of X. euvesicatoria pv. euvesicatoria; Xee (NCPPB2968, NCPPB2573), X.vesicatoria; Xv (NCPPB422, NCPPB1332, NCPPB1421), X. hortorum pv. gardneri; Xhg (NCPPB881, NCPPB4323) and X. euvesicatoria pv. perforans; Xep (NCPPB4322, NCPPB4321) and relative species; X. oryzae pv. oryzae (Xoo) X. oryzae pv. oryzicola (Xoc) and X. citri (Xcc) were used. Equal ratio mixture DNA of Xee, Xep, Xv and Xhg = Positive control (Pos). Non-template control (NTC) and M = 1 kb DNA ladder (ACTGeneTM, USA)
Detection of Xanthomonas euvesicatoria pv. perforans in tomato seed lots
Artificially inoculated seeds with bacterial cell suspensions of 1.05 x 104 to 101 CFU/ml were prepared. DNA was extracted from each seed with 5 replications of each bacterial concentration and tested. The singleplex PCR with the universal primer of 16S was done and results showed a 1,500 bp DNA band from all inoculated seeds indicating the presence of bacteria (Figure 3A). Singleplex PCR with Hpaf-F/Hpaf-R showed the highest sensitivity at 101 CFU/ml of Xep cell suspension (Figure 3B).
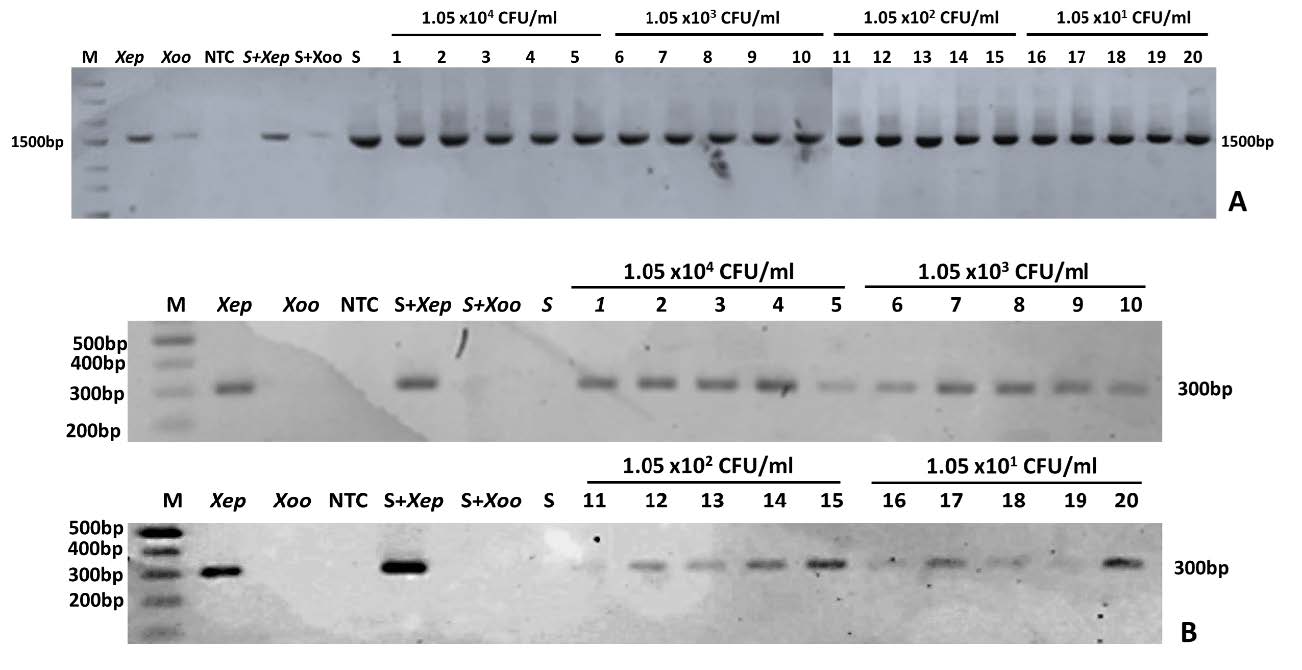
Figure 3. Xanthomonas euvesicatoria pv. perforans detection from artificially inoculated tomato seed with cell concentrations from 1.05 x 104 - 101 CFU/ml using singleplex PCR with 16S universal primer (A) and singleplex specific primer (Hpaf-F/Hpaf-R) (B); M: 100 bp DNA ladder (ACTGeneTM, USA); Xep: X. euvesicatoria pv. perforans ; Xoo: X. oryzae pv. oryzae; NTC: non-template control; S+Xep: tomato seed added with a Xep cell suspension; S+Xoo: tomato seed added with a Xoo cell suspension; S: healthy tomato seed and each tomato seed extract in various Xep cell concentrations of 1.05 x 104 CFU/ml, 1.05 x 103 CFU/ml, 1.05 x 102 CFU/ml and 1.05 x 101 CFU/ml
Similarly, the presence of bacteria in contaminated tomato seed lots was determined using 16S PCR amplification (Figure 4A). Xep-specific amplification showed the highest sensitivity at seed lot infected with 1.05 x 101 CFU/ml in all 3 replications (Figure 4B).

Figure 4. Xanthomonas euvesicatoria pv. perforans detection in 0.05% contaminated seed lot (1/2,000 seeds) with various cell concentrations of X. euvesicatoria pv. perforans from 1.05 x 104 - 101 CFU/ml by PCR method with 16S universal primer (A) and singleplex specific primer (Hpaf-F/Hpaf-R) (B); M: 1 kp DNA Ladder (ACTGeneTM, USA); Xep: X. euvesicatoria pv. perforans ; Xoo: X. oryzae pv. oryzae; NTC: non-template control; S+Xep: tomato seed added with Xep cells suspension; S+Xoo: tomato seed added with Xoo cells suspension; S: tomato seed and 3 replications of inoculated tomato seeds.
DISCUSSION
PCR based methods: conventional PCR, real-time PCR are sensitive methods and have been used to detect pathogens in many environments such as water, plant and insect (Chomvarin et al., 2017; Nguyen & Chen, 2017; Traiyasut et al., 2016). Detection methods of BLS pathogens in tomato and pepper were mainly based on PCR and real-time PCR (Koenraadt et al., 2009, Araujo et al., 2012, Osdaghi et al., 2017, Pečenka et al., 2020). They successfully detected many Xanthomonas spp. and several strains. However, the detection of X. euvesicatoria pv. perforans (Xep) from tomato products using established Bs-XpF/Bs-XpR primers (Koenraadt et al., 2009; European and Mediterranean Plant Protection Organization (EPPO) (EPPO, 2013)) in unclassified regions such as Taiwan and Thailand were shown irregularly (Osdaghi et al., 2021). In this study, primers HprF-f/HprF-r (Ning 2012) that reportedly detected Xep strain from Thailand were used. Singleplex and multiplex PCR (coupling with Xhg strain) detection were tested with or without two PCR additives (DMSO and betaine). In this experiment, a single experiment was done for investigating the potential of improving PCR amplification by adding the PCR additives which demonstrated a noticeable difference. During infection, X. vesicatoria (Xv) moved to the ovaries of tomato flowers and could be detected in the seeds (Bashan and Okon 1986). X. axonopodis pv. vesicatoria (Xav) was also detected in the inner and outer seed coats (Sharma and Agrawal, 2014). In this study, tomato seeds were artificially inoculated with Xep cell suspension by soaking and vacuuming the seeds to introduce bacteria into the seeds and eventually inside the seed coat. This method is often used to produce artificially inoculated seeds (Krttzman 1991, Hadas et al., 2005). The results showed that singleplex PCR with a combination of both additives elevated the sensitivity of Xep detection in artificially inoculated tomato seeds. Though additives showed positive effects on the sensitivity of multiplex PCR, non-specific amplification from Xep primers was observed. DMSO and betaine aid in the release of the secondary structure of the DNA, and the delay in the reannealing of the DNA template, resulting in the promotion of primer annealing and the reduction of nonspecific DNA amplification (Karunanathie et al., 2022). Varadharajan et al., (2021) reported that the addition of 5% DMSO increased the success rate of ITS2 amplification in 50 plant species from 43 genera and 29 families. However, it should be noted that the effects of additives required verification of the working DNA polymerase. Under other DNA polymerases, adverse effects of Xep amplification were observed with PCR additive supplements (data not shown). The composition of the PCR buffer, such as the types and strength of the salt, could affect the outcome.
Multiple TaqMan real-time PCR for all 4 species of bacterial leaf spot pathogens showed sensitivity detection at 105 CFU/ml (Strayer et al., 2016). PMA-qPCR for the detection of viable cells of four Xanthomonas spp. could detect seed samples spiked with 100 CFU/ml and ≥ 75 CFU/ml at a detection rate of 75 -100% and 50 -75%, respectively (Wang et al., 2022). This study tested infected seeds at 0.05% (1/2,000 seed) with initial infection at 101 CFU/ml. The maximum subsample of 10,000 seeds was recommended by the International Seed Federation (ISF) (http://www.worldseed.org/isf/ishi_vegetable.html). However, the minimum subsample size is 2,000 x 5 seeds for highly contaminated saprophytic bacteria seeds (Scortichini et al., 2013). The small subsample size gave a high chance for pathogen detection. In contamination at 0.01% or 1 infected seed in 10,000 seeds, 1 infected seed will be placed in 1 of 5 subsamples of 2,000 seeds (0.05%), which results from this work gave a high-sensitivity detection limit at 101 CFU/ml. However, it is noteworthy that in instances where the target pathogen exists in low copy numbers, a faint band may be observed.
The sigleplex PCR method, incorporating bacterial enhancement and improved PCR amplification by adding PCR additive developed in this study contributed the high potential for Xep detection with a low cost from conventional PCR-based methods and high sensitivity in contaminated seed samples making it a cost-effective and sensitive alternative to other detection methods. However, limitations such as the inability to provide quantitative information and potential ambiguity in results with low target amounts are present. The choice of Taq DNA polymerase and optimization of reaction conditions are influential factors. Future research should focus on developing multiplex primer sets for detecting multiple pathogens to enhance the assay's performance.
ACKNOWLEDGMENTS
The laboratory facilities and funding were supported by the Center of Excellence on Agricultural Biotechnology (AG-BIO/PERDO-CHE) under the project “Development standard detection method for Xanthomonas vesicatory” project no. AG-BIO/61-003-001 and Center for Agricultural Biotechnology, Kasetsart University.
AUTHOR CONTRIBUTIONS
Kanchanaphon Sawangchaitham and Parichart Burns assisted in conducting the experiments, performed the statistical analysis and data visualization and wrote the manuscript. Jutatape Watcharachaiyakup and Wichai Kositratana designed and conducted all of the experiments and wrote the manuscript. All authors have read and approved of the final manuscript.
CONFLICT OF INTEREST
The authors declare that they hold no competing interests.
REFERENCES
Araujo, E. R., Costa, J., Ferreira, M., and Quezado‐Duval, A. 2012. Simultaneous detection and identification of the Xanthomonas species complex associated with tomato bacterial spot using species‐specific primers and multiplex PCR. Journal of Applied Microbiology. 113: 1479-1490.
Chomvarin, C., Wongboot, W., Tirapattanun, A., Kanthawong, S., Wongwajana, S., Tongpim, S., and Boonyanugomol, W. 2017. Detection of Helicobacter pylori in aquatic environments and drinking waters in northeastern Thailand. Chiang Mai Journal of Science. 44: 731-741.
Constantin, E., Cleenwerck, I., Maes, M., Baeyen, S., Van Malderghem, C., De Vos, P., and Cottyn, B. 2016. Genetic characterization of strains named as Xanthomonas axonopodis pv. dieffenbachiae leads to a taxonomic revision of the X. axonopodis species complex. Plant Pathology. 65: 792-806.
Demeke, T. and Adams, R. P. 1992. The effects of plant polysaccharides and buffer additives on PCR. Biotechniques. 12: 332-334.
Dutta, B., Gitaitis, R., Sanders, H., Booth, C., Smith, S., and Langston Jr, D. 2014. Role of blossom colonization in pepper seed infestation by Xanthomonas euvesicatoria. Phytopathology. 104: 232-239.
EPPO. 2013. Xanthomonas spp.(Xanthomonas euvesicatoria, Xanthomonas gardneri, Xanthomonas perforans, Xanthomonas vesicatoria) causing bacterial spot of tomato and sweet pepper. EPPO Bulletin. 43: 7-20.
Hadas, R., Kritzman, G., Klietman, F., Gefen, T., and Manulis, S. 2005. Comparison of extraction procedures and determination of the detection threshold for Clavibacter michiganensis ssp. michiganensis in tomato seeds. Plant Pathology. 54: 643-649.
Hills, P., and Van Staden, J. 2002. An improved DNA extraction procedure for plant tissues with a high phenolic content. South African Journal of Botany. 68: 549-550.
Japelaghi, R. H., Haddad, R., and Garoosi, G.-A. 2011. Rapid and efficient isolation of high quality nucleic acids from plant tissues rich in polyphenols and polysaccharides. Molecular Biotechnology. 49: 129-137.
Jones, J., Minsavage, G., Stall, R., Kelly, R., and Bouzar, H. 1993. Genetic analysis of a DNA region involved in expression of two epitopes associated with lipopolysaccharide in Xanthomonas campestris pv. vesicatoria. Phytopathology. 83: 551-567.
Koenraadt, H., van Betteray, B., Germain, R., Hiddink, G., Jones, J. B., and Oosterhof, J. 2009. Development of specific primers for the molecular detection of bacterial spot of pepper and tomato. Acta Horticulturae. 808: 99-102
Leite Jr, R., Jones, J., Somodi, G., Minsavage, G., and Stall, R. 1995. Detection of Xanthomonas campestris pv. vesicatoria associated with pepper and tomato seed by DNA amplification. Plant Disease. 79: 917-922.
Lue, Y., Deng, W., Wu, Y., Cheng, A., Hsu, S., and Tzeng, K. 2010. Characterization of Xanthomonas associated with bacterial spot of tomato and pepper in Taiwan. Plant Pathology Bulletin. 19: 181-190.
Minsavage, G., Thompson, C., Hopkins, D., Leite, R., and Stall, R. 1994. Development of a polymerase chain reaction protocol for detection of Xylella fastidiosa in plant tissue. Phytopathology. 84: 456-461.
Morinière, L., Burlet, A., Rosenthal, E. R., Nesme, X., Portier, P., Bull, C. T., Lavire, C., Fischer-Le Saux, M., and Bertolla, F. 2020. Clarifying the taxonomy of the causal agent of bacterial leaf spot of lettuce through a polyphasic approach reveals that Xanthomonas cynarae Trébaol et al. 2000 emend. Timilsina et al. 2019 is a later heterotypic synonym of Xanthomonas hortorum Vauterin et al. 1995. Systematic and Applied Microbiology. 43: 126087.
Nguyen, P. T., and Chen, Y. C. 2017. Simultaneous detection of two viroids infecting grapevines in Taiwan by multiplex RT-PCR. Chiang Mai Journal of Science. 44: 721-730.
Ning, F. 2012. Identification and detection of Xanthomonas perforans by the polymerase chain reaction technique and characterization of X. perforans strains in Taiwan by DNA polymorphism. MSc thesis, National Chung Hsing University. Taiwan.
Osdaghi, E., Taghavi, S. M., Hamzehzarghani, H., Fazliarab, A., and Lamichhane, J. R. 2017. Monitoring the occurrence of tomato bacterial spot and range of the causal agent Xanthomonas perforans in Iran. Plant Pathology. 66: 990-1002.
Pečenka, J., Kocanová, M., Baranek, M., Gazdik, F., Ragasova, L., Peňázová, E., Čechová, J., Beran, P., and Eichmeier, A. 2020. Species-specific PCR primers for the detection of poorly distinguishable Xanthomonas euvesicatoria. Crop Protection. 127: 104978.
Roach, R., Mann, R., Gambley, C. G., Shivas, R. G., and Rodoni, B. 2018. Identification of Xanthomonas species associated with bacterial leaf spot of tomato, capsicum and chilli crops in eastern Australia. European Journal of Plant Pathology. 150: 595-608.
Ryan, R. P., Vorhölter, F.-J., Potnis, N., Jones, J. B., Van Sluys, M.-A., Bogdanove, A. J., and Dow, J. M. 2011. Pathogenomics of Xanthomonas: understanding bacterium–plant interactions. Nature Reviews: Microbiology. 9: 344-355.
Sambrook, J., and Russell, D. W. 2001. Molecular Cloning: a Laboratory Manual 3rd Edition (3 ed., Vol. 2). CSHL Press, New York, USA.
Scortichini, M., Stefani, E., Elphinstone, J., and Bergsma Vlami, M. 2013. PM 7/110 (1) Xanthomonas spp.(Xanthomonas euvesicatoria, Xanthomonas gardneri, Xanthomonas perforans, Xanthomonas vesicatoria) causing bacterial spot of tomato and sweet pepper. EPPO Bulletin. 43: 7-20.
Sheu, D.-S., Wang, Y.-T., and Lee, C.-Y. 2000. Rapid detection of polyhydroxyalkanoate-accumulating bacteria isolated from the environment by colony PCR. Microbiology. 146: 2019-2025.
Sitthitanasin, S., Korakngam, C., Kanhayart, T., Watcharachaiyakup, J., Kositcharoenkul, N., Patarapuwadol, S., and Kositratana, W. 2020. Characterization of Xanthomonas causing of bacterial leaf spot of tomato and pepper in Thailand. Thai Agricultural Research Journal. 38: 80-89.
Sriwilai, B. 1994. Classification and detection of Xanthomonas campestris pv. vesicatoria (Doidge) Dye a causal agent of bacterial spot of tomato. MSc Thesis, Khon Kaen University. Khon Kaen, Thailand.
Traiyasut, P., Mookhploy, W., Kimura, K., Yoshiyama, M., Khongphinitbunjong, K., and Chantawannakul, P. 2016. First detection of honey bee viruses in wax moth. Chiang Mai Journal of Science. 43: 695-698.
Utami, D., Meale, S. J., and Young, A. J. 2022. A pan-global study of bacterial leaf spot of chilli caused by Xanthomonas spp. Plants. 11: 2291.
Varadharajan, B., Parani, M., and Boatwright, J. S. 2021. DMSO and betaine significantly enhance the PCR amplification of ITS2 DNA barcodes from plants. Genome. 64: 165-171.
van Pelt-Verkuil, E., Van Belkum, A. and Hays, J.P.2008. Principles and Technical Aspects of PCR Amplification. Springer Dordrecht,
Walcott, R. R. 2003. Detection of seedborne pathogens. HortTechnology. 13: 40-47.
Wang, H., Wagnon, R., Moreno, D., Timilsina, S., Jones, J., Vallad, G., and Turechek, W. W. 2022. A long-amplicon viability-qPCR test for quantifying living pathogens that cause bacterial spot in tomato seed. Plant Disease. 106: 1474-1485.
Wang, T.-T., Yin, K.-Q., and Zhang, L.-P. 2018. Betaine improves the PCR amplification of rice GC-rich DNA sequence. Biotechnology Bulletin. 34: 80-86.
Weisburg, W. G., Barns, S. M., Pelletier, D. A., and Lane, D. J. 1991. 16S ribosomal DNA amplification for phylogenetic study. Journal of Bacteriology. 173: 697-703.
OPEN access freely available online
Natural and Life Sciences Communications
Chiang Mai University, Thailand. https://cmuj.cmu.ac.th
Jutatape Watcharachaiyakup1, 2, *, Kanchanaphon Sawangchaitham1, 2, Parichart Burns3 and Wichai Kositratana1, 2, *
1 Center for Agricultural Biotechnology, Kasetsart University, Kamphaeng Sean Campus, Nakhon Pathom 73140, Thailand.
2 Center of Excellence on Agricultural Biotechnology (AG-BIO/PERDO-CHE), Bangkok 10900, Thailand.
3 National Center for Genetic Engineering and Biotechnology, National Science and Technology Development Agency, Pathum Thani 12120, Thailand.
Corresponding author: Jutatape Watcharachaiyakup E-mail: jutatape.w@ku.th,
Wichai Kositratana E-mail: agrwck@ku.ac.th
Total Article Views
Editor: Tonapha Pusadee,
Chiang Mai University, Thailand
Article history:
Received: April 10, 2023;
Revised: June 22, 2023;
Accepted: June 26, 2023;
Published online: July 10, 2023

